7QX3
 
 | | Structure of the transaminase TR2E2 with EOS | | Descriptor: | 2-azanylethyl hydrogen sulfate, Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7QX0
 
 | | Transaminase Structure of Plurienzyme (Tr2E2) in complex with PLP | | Descriptor: | Aminotransferase TR2, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QYG
 
 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
5UJ7
 
 | | Structure of the active form of human Origin Recognition Complex ATPase motor module, complex subunitS 1, 4, 5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, Elkayam, E, On, K.F, Joshua-Tor, L. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
7QFV
 
 | | Crystal structure of KLK6 in complex with compound 17a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Ser(Z)-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7QFT
 
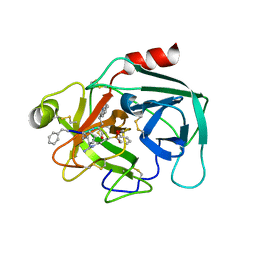 | | Crystal structure of KLK6 in complex with compound 16a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Cha-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
1JFV
 
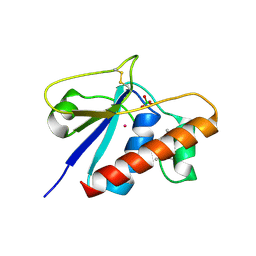 | | X-Ray Structure of oxidised C10S, C15A arsenate reductase from pI258 | | Descriptor: | PERCHLORATE ION, POTASSIUM ION, arsenate reductase | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
1JH5
 
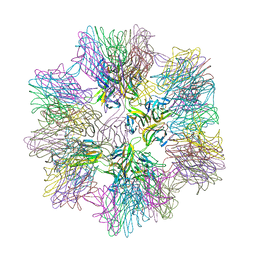 | | Crystal Structure of sTALL-1 of TNF family ligand | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Liu, Y, Xu, L, Opalka, N, Shu, H.-B, Zhang, G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands.
Cell(Cambridge,Mass.), 108, 2002
|
|
4NFV
 
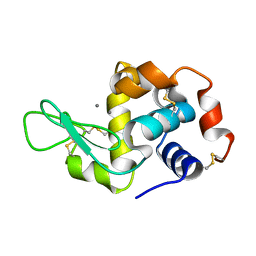 | | Previously de-ionized HEW lysozyme batch crystallized in 1.1 M MnCl2 | | Descriptor: | CHLORIDE ION, Lysozyme C, MANGANESE (II) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1OAJ
 
 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castelli, S, Falconi, M, Parrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-27 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
1NM8
 
 | | Structure of Human Carnitine Acetyltransferase: Molecular Basis for Fatty Acyl Transfer | | Descriptor: | Carnitine O-acetyltransferase | | Authors: | Wu, D, Govindasamy, L, Lian, W, Gu, Y, Kukar, T, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Human Carnitine Acetyltransferase. Molecular Basis for Fatty Acyl Transfer
J.Biol.Chem., 278, 2003
|
|
7RJ3
 
 | | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycogen accumulation regulator GarA | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis.
To Be Published
|
|
5UKB
 
 | | VSV N PROTEIN IN COMPLEX WITH INHIBITORY NANOBODY 1004 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Nucleocapsid, RNA (45-MER) | | Authors: | Hanke, L, Knockenhauer, K.E, Ploegh, H.L, Schwartz, T.U. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (5.473 Å) | | Cite: | Vesicular stomatitis virus N protein-specific single-domain antibody fragments inhibit replication.
EMBO Rep., 18, 2017
|
|
7RJJ
 
 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5UOI
 
 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | Descriptor: | HHH_rd1_0142 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UZ9
 
 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
1JXG
 
 | | The 1.6 A Resolution Crystal Structure of a Mutant Poplar Plastocyanin Bearing a 21-25 Engeneered Disulfide Bridge | | Descriptor: | COPPER (I) ION, PLASTOCYANIN A, SULFATE ION | | Authors: | Milani, M, Andolfi, L, Cannistraro, S, Verbeet, M.P, Bolognesi, M. | | Deposit date: | 2001-09-07 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution crystal structure of a mutant plastocyanin bearing a 21-25 engineered disulfide bridge.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
5V6Z
 
 | |
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7REF
 
 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7RBQ
 
 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
1JBA
 
 | | UNMYRISTOYLATED GCAP-2 WITH THREE CALCIUM IONS BOUND | | Descriptor: | CALCIUM ION, PROTEIN (GUANYLATE CYCLASE ACTIVATING PROTEIN 2) | | Authors: | Ames, J.B, Dizhoor, A.M, Ikura, M, Palczewski, K, Stryer, L. | | Deposit date: | 1999-04-03 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases.
J.Biol.Chem., 274, 1999
|
|
1NHU
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
