6YXD
 
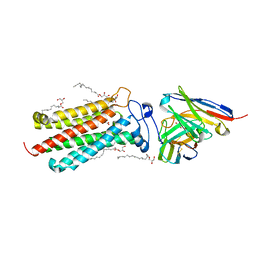 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXF
 
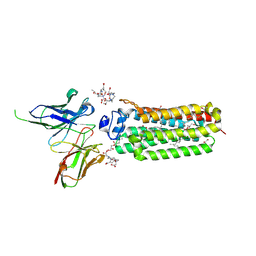 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXH
 
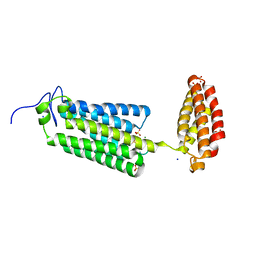 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6ZCU
 
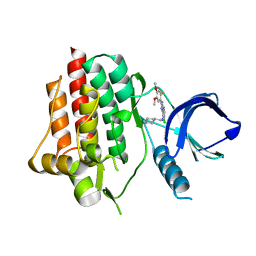 | | syk in complex with 57262_SYKB-AZ13344324-2 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCP
 
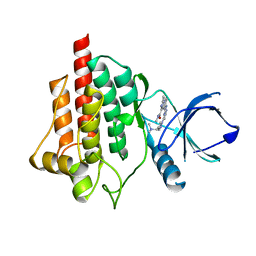 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | 7-[[4-[4-[(dimethylamino)methyl]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 4
To Be Published
|
|
6ZCS
 
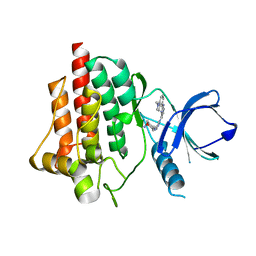 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 3 | | Descriptor: | (3~{R},4~{R})-~{N}4-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]oxane-3,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 3
To Be Published
|
|
6ZC0
 
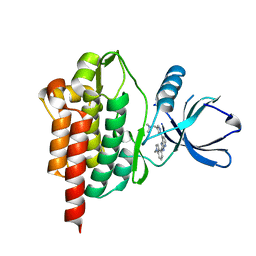 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},~{N}-dimethyl-1-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]methanamine | | Authors: | Read, J.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCY
 
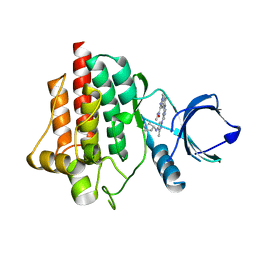 | | SYK Kinase domain in complex with diamine inhibitor 16 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-pyrrolidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 16
To Be Published
|
|
6ZCX
 
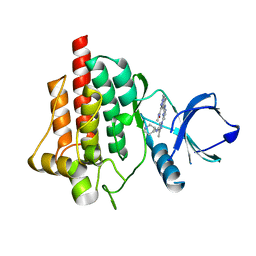 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 18 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-piperidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 18
To Be Published
|
|
6ZCQ
 
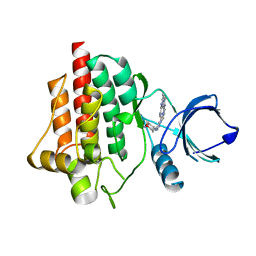 | | SYK Kinase domain in complex with diamine inhibitor 5 | | Descriptor: | 7-[[4-[4-[[(3~{R},4~{R})-3-azanyloxan-4-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | SYK Kinase domain in complex with diamine inhibitor 5
To Be Published
|
|
6ZCR
 
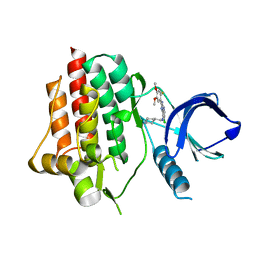 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 7 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 7
To Be Published
|
|
1NK3
 
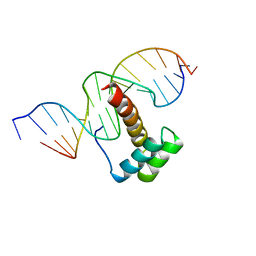 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK2
 
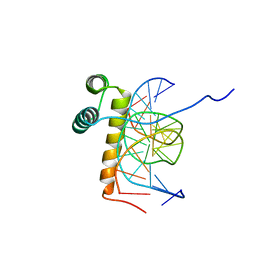 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
6OE3
 
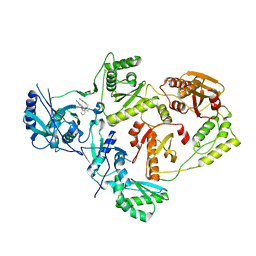 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Kudalkar, S.N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and pharmacological evaluation of a novel non-nucleoside reverse transcriptase inhibitor as a promising long acting nanoformulation for treating HIV.
Antiviral Res., 167, 2019
|
|
1TV3
 
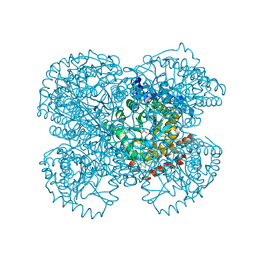 | | Crystal structure of the N-methyl-hydroxylamine MtmB complex | | Descriptor: | 5-(HYDROXY-METHYL-AMINO)-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
7ABS
 
 | | Structure of human DCLRE1C/Artemis in complex with DNA - re-evaluation of 6WO0 | | Descriptor: | DNA (5'-D(*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*GP*CP*T)-3'), Protein artemis, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7A6G
 
 | |
7AAL
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1, G258A mutant | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAM
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 bound to the CTH domain of the phosphatase LYP | | Descriptor: | GLYCEROL, Proline-serine-threonine phosphatase-interacting protein 1, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAN
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AFS
 
 | | The structure of Artemis variant D37A | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7B2X
 
 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7APV
 
 | | Structure of Artemis/DCLRE1C/SNM1C in complex with Ceftriaxone | | Descriptor: | 1,2-ETHANEDIOL, Ceftriaxone, NICKEL (II) ION, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AF1
 
 | | The structure of Artemis/SNM1C/DCLRE1C with 2 Zinc ions | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
