2UYQ
 
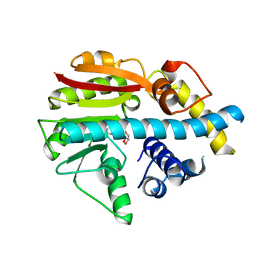 | | Crystal structure of ML2640c from Mycobacterium leprae in complex with S-adenosylmethionine | | Descriptor: | HYPOTHETICAL PROTEIN ML2640, S-ADENOSYLMETHIONINE | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
7QGE
 
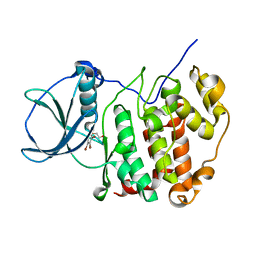 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6,7,8-TETRABROMOBENZOTRIAZOLE (TBBt) AT PH 8.5 | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGD
 
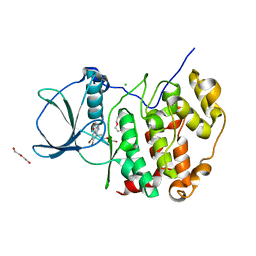 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 8.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGB
 
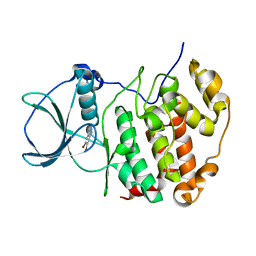 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 6.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, Casein kinase II subunit alpha | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
6S68
 
 | | Structure of the Fluorescent Protein AausFP2 from Aequorea cf. australis at pH 7.6 | | Descriptor: | Aequorea cf. australis fluorescent protein 2 (AausFP2) | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
7QGG
 
 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
4YEH
 
 | | Crystal structure of Mg2+ ion containing hemopexin fold from Kabuli chana (chickpea white) at 2.45A resolution reveals a structural basis of metal ion transport | | Descriptor: | Lectin, MAGNESIUM ION | | Authors: | Kumar, S, Singh, A, Yamini, S, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Mg(2+) Containing Hemopexin-Fold Protein from Kabuli Chana (Chickpea-White, CW-25) at 2.45 angstrom Resolution Reveals Its Metal Ion Transport Property
Protein J., 34, 2015
|
|
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
1NOG
 
 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
2AKK
 
 | | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris | | Descriptor: | phnA-like protein | | Authors: | Wu, B, Yee, A, Ramelot, T.A, Semesi, A, Lemak, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris
To be Published
|
|
4KUO
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
4YN8
 
 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | Descriptor: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | Authors: | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2015-03-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3ZF6
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A D110C S168C mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
2KA3
 
 | | Structure of EMILIN-1 C1Q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Corazza, A, Colebrooke, S.A, Cicero, D.O, Eliseo, T, Boyd, J, Doliana, R, Fogolari, F, Viglino, P, Colombatti, A, Campbell, I.D, Esposito, G. | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
J.Biomol.Nmr, 43, 2009
|
|
3ZF1
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81N mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
7OBN
 
 | | Structural investigations of a new L3 DNA ligase: structure-function analysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), DNA ligase, ... | | Authors: | Leiros, H.-K.S, Williamson, A. | | Deposit date: | 2021-04-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bacteriophage origin of some minimal ATP-dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase.
Sci Rep, 11, 2021
|
|
3ZF5
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84F mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
7OBI
 
 | | Consensus tetratricopeptide repeat protein type RV4 | | Descriptor: | CTPR-rv4, PHOSPHATE ION | | Authors: | Eapen, R.S, Perez-Riba, A, Fischer, G, Itzhaki, L.S, Hyvonen, M. | | Deposit date: | 2021-04-22 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unraveling the Mechanics of a Repeat-Protein Nanospring: From Folding of Individual Repeats to Fluctuations of the Superhelix.
Acs Nano, 16, 2022
|
|
1CA2
 
 | |
2V40
 
 | | Human Adenylosuccinate synthetase isozyme 2 in complex with GDP | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE ISOZYME 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Welin, M, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Adenylosuccinate Synthetase Isozyme 2 in Complex with Gdp
To be Published
|
|
2K4V
 
 | | Solution structure of uncharacterized protein PA1076 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium (NESG) target PaT3, Ontario Center for Structural Proteomics target PA1076 . | | Descriptor: | uncharacterized protein PA1076 | | Authors: | Gutmanas, A, Lemak, A, Fares, C, Yee, A, Semesi, A, Arrowsmith, C.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-06-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized protein PA1076 from Pseudomonas aeruginosa.
To be Published
|
|
2UZ9
 
 | | Human guanine deaminase (guaD) in complex with zinc and its product Xanthine. | | Descriptor: | GUANINE DEAMINASE, XANTHINE, ZINC ION | | Authors: | Moche, M, Welin, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Guanine Deaminase (Guad) in Complex with Zinc and its Product Xhantine
To be Published
|
|
3QVM
 
 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | Authors: | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
