1NXN
 
 | | SOLUTION STRUCTURE OF CONTRYPHAN-VN | | Descriptor: | CONTRYPHAN-VN, MAJOR FORM (CIS CONFORMER) | | Authors: | Eliseo, T, Cicero, D.O, Polticelli, F, Schinina, M.E, Massilia, G.R, Paci, M, Ascenzi, P. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-04 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cyclic peptide contryphan-Vn, a Ca2+-dependent K+ channel modulator
Biopolymers, 74, 2004
|
|
1ZZF
 
 | |
2K1Q
 
 | | NMR structure of hepatitis c virus ns3 serine protease complexed with the non-covalently bound phenethylamide inhibitor | | Descriptor: | NS3 PROTEASE, PHENETHYLAMIDE, ZINC ION | | Authors: | Eliseo, T, Gallo, M, Pennestri, M, Bazzo, R, Cicero, D.O. | | Deposit date: | 2008-03-13 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Binding of a noncovalent inhibitor exploiting the S' region stabilizes the hepatitis C virus NS3 protease conformation in the absence of cofactor.
J.Mol.Biol., 385, 2009
|
|
2JN3
 
 | | NMR structure of cl-BABP complexed to chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Fatty acid-binding protein, liver | | Authors: | Eliseo, T, Ragona, L, Catalano, M, Assfalf, M, Paci, M, Zetta, L, Molinari, H, Cicero, D.O. | | Deposit date: | 2006-12-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic determinants of ligand binding in the ternary complex of chicken liver bile acid binding protein with two bile salts revealed by NMR
Biochemistry, 46, 2007
|
|
6FE6
 
 | | Solution structure of a last generation P2-P4 macrocyclic inhibitor | | Descriptor: | (3aR,7S,10S,12R,24aR)-7-cyclopentyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-5,8-dioxo-1,2,3,3a,5,6,7,8,11,12,20,21,22,23,24,24a-hexadecahydro-10H-9,12-methanocyclopenta[18,19][1,10,3,6]dioxadiazacyclononadecino[12,11-b]quinoline-10-carboxamide, Non-structural 3 protease, ZINC ION | | Authors: | Gallo, M, Eliseo, T, Cicero, D.O. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a last generation macrocyclic inhibitor. Hepatitis C virus NS3 protease complex: when S prime region occupancy is not enough to stabilize the protein conformation in the absence of NS4A.
To Be Published
|
|
2MNQ
 
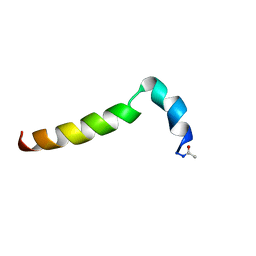 | | 1H, 13C, and 15N Chemical Shift Assignments for Thymosin alpha 1 | | Descriptor: | THYMOSIN ALPHA-1 | | Authors: | Nepravishta, R, Mandaliti, W, Eliseo, T, Sinibaldi Vallebona, P, Pica, F, Garaci, E, Paci, M. | | Deposit date: | 2014-04-09 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Thymosin alpha 1 inserts N terminus into model membranes assuming a helical conformation.
Expert Opin Biol Ther, 15 Suppl 1, 2015
|
|
2OII
 
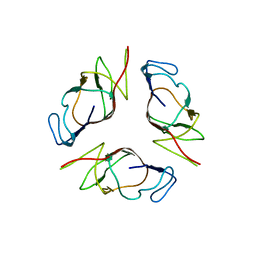 | | Structure of EMILIN-1 C1q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Colebrooke, S.A, Corazza, A, Cicero, D.O, Eliseo, T, Viglino, P, Campbell, I.D, Colombatti, A, Esposito, G. | | Deposit date: | 2007-01-11 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of EMILIN-1
To be Published
|
|
1R8P
 
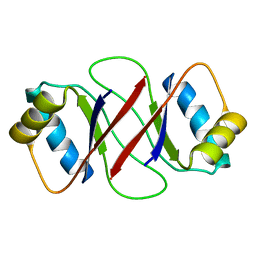 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1ZRY
 
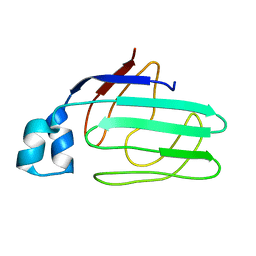 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
2K6L
 
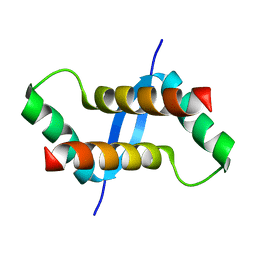 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
2KA3
 
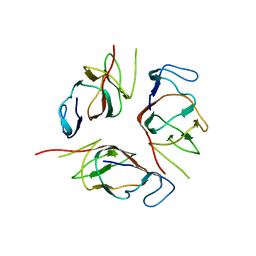 | | Structure of EMILIN-1 C1Q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Corazza, A, Colebrooke, S.A, Cicero, D.O, Eliseo, T, Boyd, J, Doliana, R, Fogolari, F, Viglino, P, Colombatti, A, Campbell, I.D, Esposito, G. | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
J.Biomol.Nmr, 43, 2009
|
|
