5LB8
 
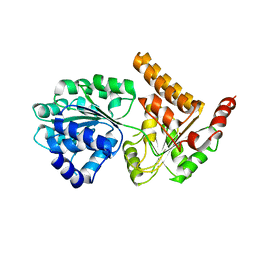 | | Crystal structure of human RECQL5 helicase APO form. | | Descriptor: | ATP-dependent DNA helicase Q5, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
5LB5
 
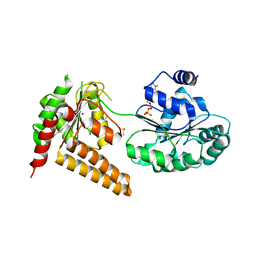 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg (tricilinc form). | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, DIMETHYL SULFOXIDE, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
6Z83
 
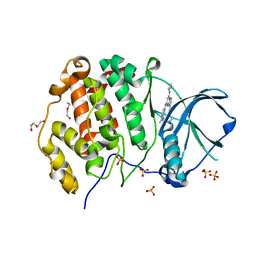 | | CK2 alpha bound to chemical probe SGC-CK2-1 | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
5LEV
 
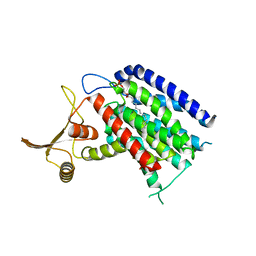 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | Descriptor: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5LXD
 
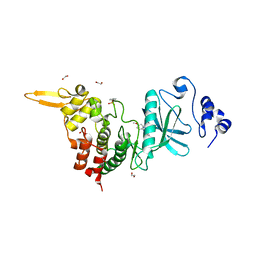 | | Crystal structure of DYRK2 in complex with EHT 1610 (compound 2) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
6Z84
 
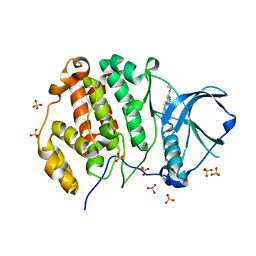 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
5LWN
 
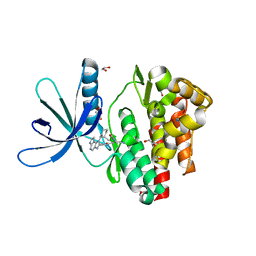 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | Descriptor: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
6TCX
 
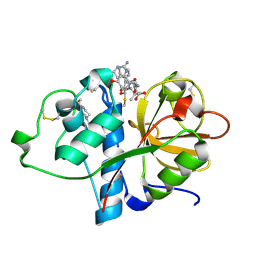 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
4RCM
 
 | | Crystal structure of the Pho92 YTH domain in complex with m6A | | Descriptor: | Methylated RNA-binding protein 1, RNA (5'-R(*UP*G)-D(*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
5JJZ
 
 | | Chromo domain of human Chromodomain Protein, Y-Like 2 | | Descriptor: | Chromodomain Y-like protein 2, LYS-LYS-LYS-ALA-ARG-MLY-SER-ALA-GLY-ALA-ALA-LYS-TYR | | Authors: | DONG, A, DOMBROVSKI, L, LOPPNAU, P, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of CDYL2 domain of human CDYL2 protein
to be published
|
|
4RCI
 
 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
5YBJ
 
 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
4R3I
 
 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
6PI7
 
 | |
2A87
 
 | | Crystal Structure of M. tuberculosis Thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Akif, M, Suhre, K, Verma, C, Mande, S.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6V2S
 
 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2H
 
 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5YBV
 
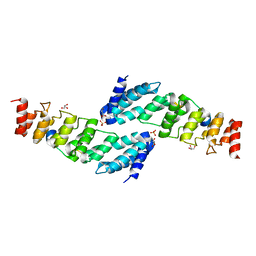 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5KE3
 
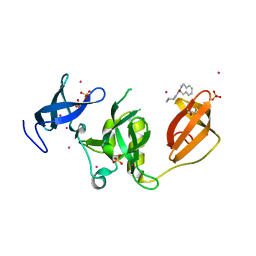 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | Descriptor: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
5YBU
 
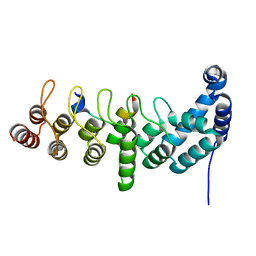 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
6V2R
 
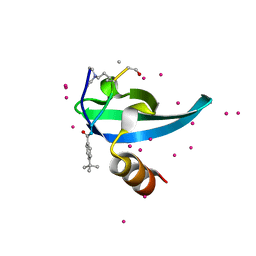 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5KE2
 
 | | Crystal structure of SETDB1 Tudor domain in complex with inhibitor XST06472A | | Descriptor: | (3~{S})-~{N}-~{tert}-butyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with inhibitor xst06472a
to be published
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
6WAU
 
 | | Complex structure of PHF19 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 19, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
