2E9O
 
 | | Structure of h-CHK1 complexed with AA582939 | | Descriptor: | 4-(6-{[(4-METHYLCYCLOHEXYL)AMINO]METHYL}-1,4-DIHYDROINDENO[1,2-C]PYRAZOL-3-YL)BENZOIC ACID, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 1,4-dihydroindeno[1,2-c]pyrazoles as a novel class of potent and selective checkpoint kinase 1 inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
2E9P
 
 | | Structure of h-CHK1 complexed with A771129 | | Descriptor: | 1-(5-CHLORO-2-METHOXYPHENYL)-3-{6-[2-(DIMETHYLAMINO)-1-METHYLETHOXY]PYRAZIN-2-YL}UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent and Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9U
 
 | | Structure of h-CHK1 complexed with A780125 | | Descriptor: | 18-CHLORO-11,12,13,14-TETRAHYDRO-1H,10H-8,4-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECIN-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2FME
 
 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5D1I
 
 | |
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
4X1K
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-alanyl-N-[(3R,4S,5S)-1-{(2S)-2-[(1R,2R)-3-{[(1S)-1-carboxy-2-phenylethyl]amino}-1-methoxy-2-methyl-3-oxopropyl]pyrrolidin-1-yl}-3-methoxy-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
4X20
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-prolyl-N-[(3R,4S,5S)-1-{(2S)-2-[(1R,2R)-3-{[(1S)-1-carboxy-2-phenylethyl]amino}-1-methoxy-2-methyl-3-oxopropyl]pyrrolidin-1-yl}-3-methoxy-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
4X1I
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-alanyl-N-[(3R,4S,5S)-3-methoxy-1-{(2S)-2-[(1R,2R)-1-methoxy-2-methyl-3-oxo-3-{[(1S)-2-phenyl-1-(1,3-thiazol-2-yl)ethyl]amino}propyl]pyrrolidin-1-yl}-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
4X1Y
 
 | |
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4Y19
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS18_alpha, FS18_beta, ... | | Authors: | Beringer, D.X, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
4Y1A
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS17_alpha, FS17_beta, ... | | Authors: | Beringer, D.X, Vivian, J.P, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
4Y3U
 
 | | The structure of phospholamban bound to the calcium pump SERCA1a | | Descriptor: | Cardiac phospholamban, POTASSIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Hurley, T.D. | | Deposit date: | 2015-02-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
J. Biol. Chem., 288, 2013
|
|
7YTU
 
 | |
7YTT
 
 | |
7YS6
 
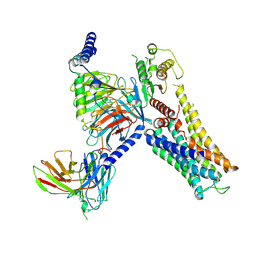 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4HXJ
 
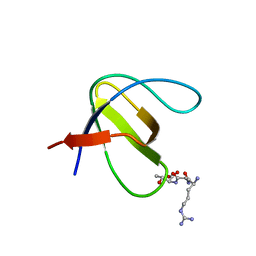 | | Crystal structure of SH3:RGT complex | | Descriptor: | C-terminal 3-mer peptide from Integrin beta-3, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Xiao, R, Meng, G. | | Deposit date: | 2012-11-11 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural framework of c-Src activation by integrin beta 3
Blood, 121, 2013
|
|
6P66
 
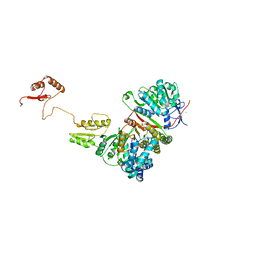 | |
6PE4
 
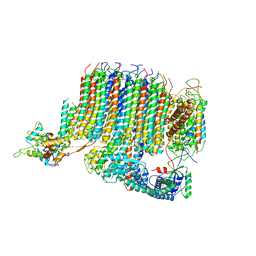 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6P4F
 
 | | Crystal structure of the XPB-Bax1-forked DNA ternary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*TP*TP*TP*GP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*GP*TP*AP*GP*GP*TP*TP*TP*CP*CP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | He, F, Hilario, E, Fan, L. | | Deposit date: | 2019-05-27 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of the XPB helicase-Bax1 nuclease complex interacting with the repair bubble DNA.
Nucleic Acids Res., 48, 2020
|
|
6PE5
 
 | | Yeast Vo motor in complex with 2 VopQ molecules | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2OCV
 
 | | Structural basis of Na+ activation mimicry in murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
