4ELO
 
 | | Ferric binding protein in apo form 1 | | Descriptor: | Iron ABC transporter, periplasmic iron-binding protein | | Authors: | Wang, Q, Liu, X.Q, Wang, X.Q. | | Deposit date: | 2012-04-11 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Crystal structure of the ferric binding protein A
To be Published
|
|
2I0N
 
 | | Structure of Dictyostelium discoideum Myosin VII SH3 domain with adjacent proline rich region | | Descriptor: | Class VII unconventional myosin | | Authors: | Wang, Q, Deloia, M.A, Kang, Y, Litchke, C, Titus, M.A, Walters, K.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SH3 domain of a M7 interacts with its C-terminal proline-rich region.
Protein Sci., 16, 2007
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
3HAJ
 
 | | Crystal structure of human PACSIN2 F-BAR domain (p212121 lattice) | | Descriptor: | CALCIUM ION, human PACSIN2 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HAH
 
 | | Crystal structure of human PACSIN1 F-BAR domain (C2 lattice) | | Descriptor: | CALCIUM ION, human PACSIN1 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HAI
 
 | | Crystal structure of human PACSIN1 F-BAR domain (P21 lattice) | | Descriptor: | CALCIUM ION, human PACSIN1 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2RFU
 
 | | Crystal structure of influenza B virus hemagglutinin in complex with LSTc receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Influenza B hemagglutinin (HA), ... | | Authors: | Wang, Q, Tian, X, Chen, X, Ma, J. | | Deposit date: | 2007-10-01 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor specificity of influenza B virus hemagglutinin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RFT
 
 | | Crystal structure of influenza B virus hemagglutinin in complex with LSTa receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Influenza B hemagglutinin (HA), ... | | Authors: | Wang, Q, Tian, X, Chen, X, Ma, J. | | Deposit date: | 2007-10-01 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor specificity of influenza B virus hemagglutinin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8UP4
 
 | | Crystal structure of Lsd18 (a flavin-dependent monooxygenase) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase Lsd18 | | Authors: | Wang, Q, Chen, X, Kim, C.Y. | | Deposit date: | 2023-10-20 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis of Sequential Enantioselective Epoxidation by a Flavin-Dependent Monooxygenase in Lasalocid A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8T3P
 
 | |
3SVR
 
 | |
3SVU
 
 | |
3SVS
 
 | |
3SVO
 
 | |
3SVN
 
 | |
7BZF
 
 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-04-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
6AEE
 
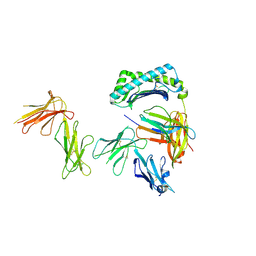 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6AED
 
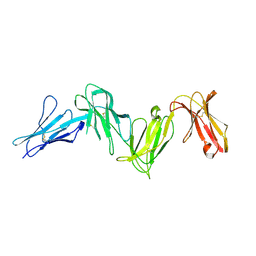 | |
8W5J
 
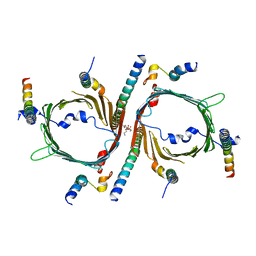 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
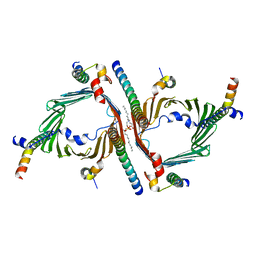 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
7XL7
 
 | |
7Y9M
 
 | | Crystal structure of P450 BM3-2F from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium
to be published
|
|
