5F6K
 
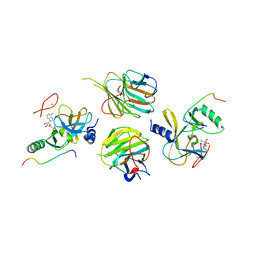 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
7DHI
 
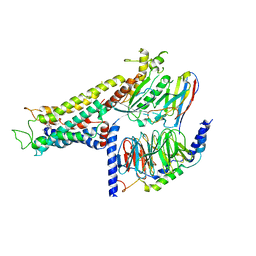 | | Cryo-EM structure of the partial agonist salbutamol-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-15 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
7DHR
 
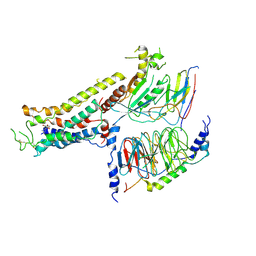 | | Cryo-EM structure of the full agonist isoprenaline-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
7CFS
 
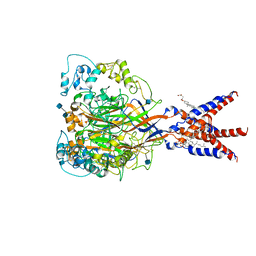 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFT
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
2L26
 
 | | Rv0899 from Mycobacterium tuberculosis contains two separated domains | | Descriptor: | Uncharacterized protein Rv0899/MT0922 | | Authors: | Shi, C, Li, J, Gao, Y, Wu, K, Huang, H, Tian, C. | | Deposit date: | 2010-08-12 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Mycobacterium tuberculosis Rv0899 Reveal a Monomeric Membrane-Anchoring Protein with Two Separate Domains
J.Mol.Biol., 2011
|
|
7S0D
 
 | |
7S0E
 
 | |
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
 | |
7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WIH
 
 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7W6L
 
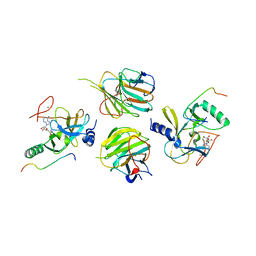 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
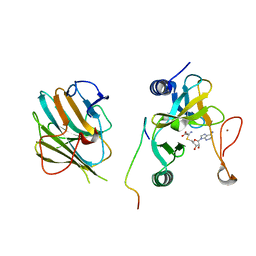 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
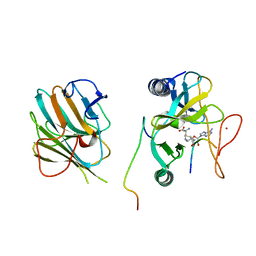 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7WKU
 
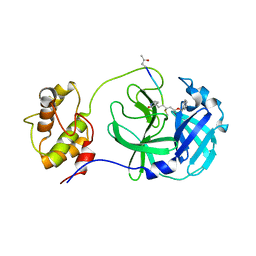 | | Structure of PDCoV Mpro in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Peptidase C30 | | Authors: | Wang, F.H, Yang, H.T. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Viruses, 14, 2022
|
|
5C7O
 
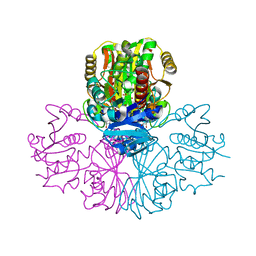 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
4UM5
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and Phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UME
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMD
 
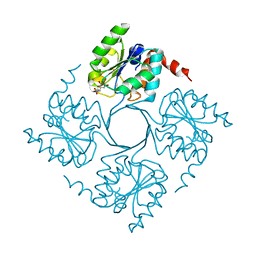 | |
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
