5E6M
 
 | | Crystal structure of human wild type GlyRS bound with tRNAGly | | Descriptor: | GLYCYL-ADENOSINE-5'-PHOSPHATE, Glycine--tRNA ligase, NICKEL (II) ION, ... | | Authors: | Xie, W, Qin, X, Deng, X, Chen, L, Liu, Y. | | Deposit date: | 2015-10-10 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.927 Å) | | Cite: | Crystal Structure of the Wild-Type Human GlyRS Bound with tRNA(Gly) in a Productive Conformation
J.Mol.Biol., 428, 2016
|
|
5BOO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
4B99
 
 | | Crystal Structure of MAPK7 (ERK5) with inhibitor | | Descriptor: | 11-cyclopentyl-2-[[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl-phenyl]amino]-5-methyl-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MITOGEN-ACTIVATED PROTEIN KINASE 7 | | Authors: | Elkins, J.M, Wang, J, Vollmar, M, Mahajan, P, Savitsky, P, Deng, X, Gray, N.S, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-03 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Crystal Structure of Erk5 (Mapk7) in Complex with a Specific Inhibitor.
J.Med.Chem., 56, 2013
|
|
4CC6
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
6W7V
 
 | | Structure of rabbit actin in complex with truncated analog of Mycalolide B | | Descriptor: | (1E,3R,4R,5S,6R,9S,10S,12S)-12-[(4-aminobutanoyl)oxy]-1-[ethyl(formyl)amino]-4,10-dimethoxy-3,5,9,13-tetramethyltetradec-1-en-6-yl (2R)-oxolane-2-carboxylate, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Allingham, J.S, Deng, X, Trofimova, D. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Truncated Actin-Targeting Macrolide Derivative Blocks Cancer Cell Motility and Invasion of Extracellular Matrix.
J.Am.Chem.Soc., 143, 2021
|
|
6E0B
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase C276F mutant bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Tomchick, D.R, Phillips, M.A, Deng, X. | | Deposit date: | 2018-07-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification and Mechanistic Understanding of Dihydroorotate Dehydrogenase Point Mutations in Plasmodium falciparum that Confer in Vitro Resistance to the Clinical Candidate DSM265.
ACS Infect Dis, 5, 2019
|
|
4HQM
 
 | | The crystal structure of QsrR-menadione complex | | Descriptor: | 2-methylnaphthalene-1,4-diol, QsrR protein | | Authors: | Ji, Q, Zhang, L, Jones, M.B, Sun, F, Deng, X, Liang, H, Brugarolas, P, Gao, N, Peterson, S.N, Lan, L, Bae, T, He, C. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GXO
 
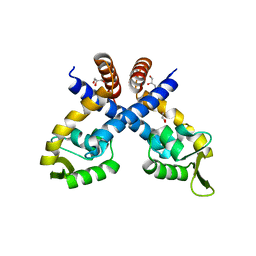 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8W2F
 
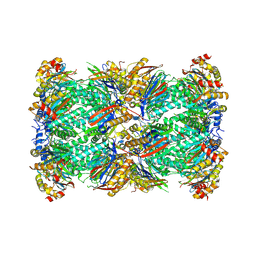 | | Plasmodium falciparum 20S proteasome bound to an inhibitor | | Descriptor: | (3S)-1-[(2-fluoroethoxy)acetyl]-N-{[(4P)-4-(6-methylpyridin-3-yl)-1,3-thiazol-2-yl]methyl}piperidine-3-carboxamide, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Han, Y, Deng, X, Ray, S, Chen, Z, Phillips, M. | | Deposit date: | 2024-02-20 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of potent and reversible piperidine carboxamides that are species-selective orally active proteasome inhibitors to treat malaria.
Cell Chem Biol, 31, 2024
|
|
6IRE
 
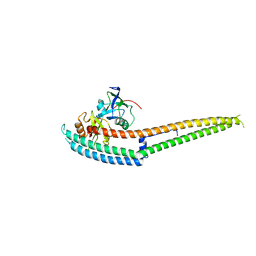 | | Complex structure of INAD PDZ45 and NORPA CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, Inactivation-no-after-potential D protein | | Authors: | Ye, F, Li, J, Deng, X, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
7P0K
 
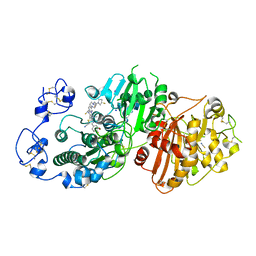 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
6RIA
 
 | |
4KQE
 
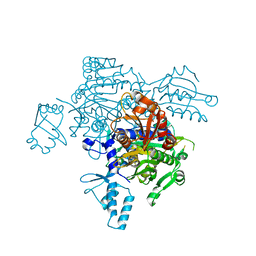 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
4DNW
 
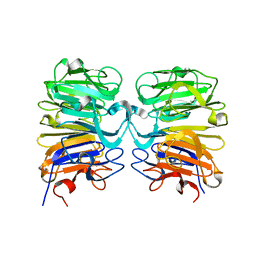 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
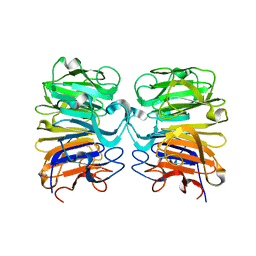 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2017-06-24 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
7Y8F
 
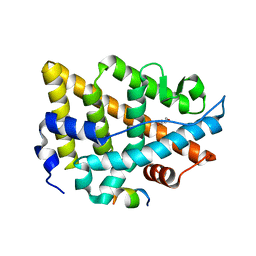 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with an Inhibitor 30o and GRIP Peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | Authors: | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-guided identification of novel dual-targeting estrogen receptor alpha degraders with aromatase inhibitory activity for the treatment of endocrine-resistant breast cancer.
Eur.J.Med.Chem., 253, 2023
|
|
