7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3L
 
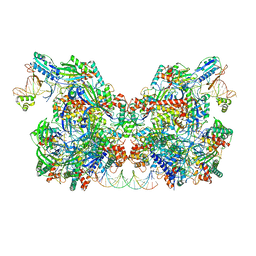 | | Cryo-EM structure of Csy-AcrIF24-DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
5G04
 
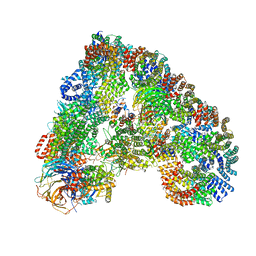 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5G05
 
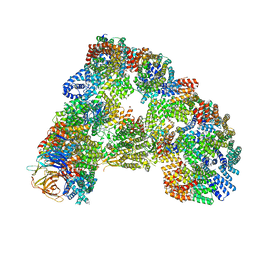 | | Cryo-EM structure of combined apo phosphorylated APC | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
1S9X
 
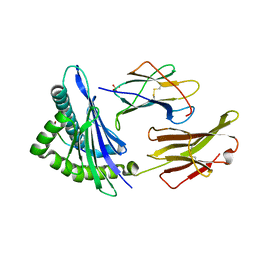 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQA, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9Y
 
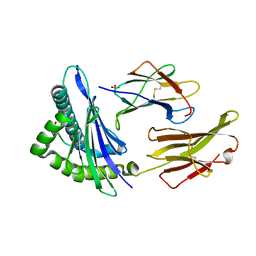 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9W
 
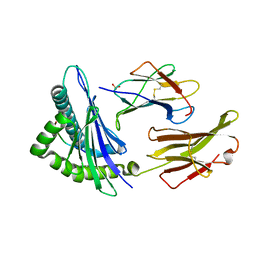 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1TFP
 
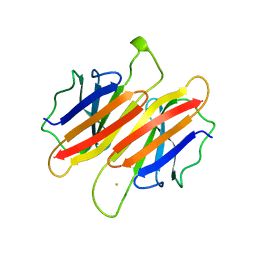 | | TRANSTHYRETIN (FORMERLY KNOWN AS PREALBUMIN) | | Descriptor: | SULFATE ION, TRANSTHYRETIN | | Authors: | Sunde, M, Richardson, S.J, Chang, L, Pettersson, T.M, Schreiber, G, Blake, C.C.F. | | Deposit date: | 1996-01-05 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of transthyretin from chicken.
Eur.J.Biochem., 236, 1996
|
|
7KC2
 
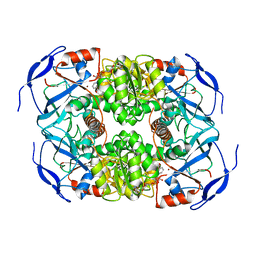 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCB
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NAD+ and Trifluoroethanol | | Descriptor: | ADH1 isoform 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCQ
 
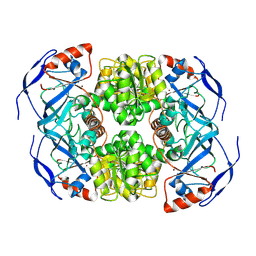 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Open Form of Apoenzyme | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KJY
 
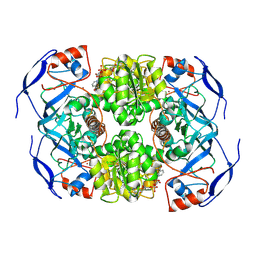 | | Symmetry in Yeast Alcohol Dehydrogenase 1 - Open Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
9CGX
 
 | | Alzheimer's Disease Seeded 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
9CGZ
 
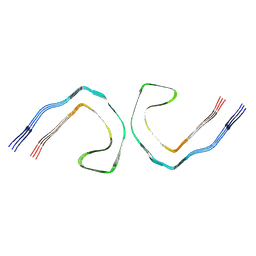 | | Alzheimer's Disease Seeded Mixed 0N4R and 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
1A90
 
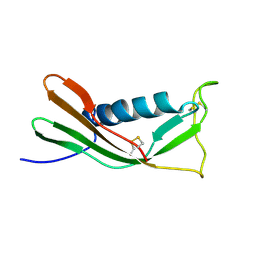 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1A67
 
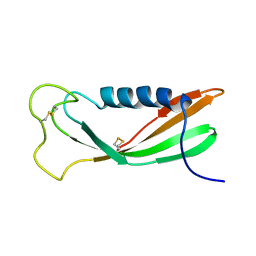 | | CHICKEN EGG WHITE CYSTATIN WILDTYPE, NMR, 16 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1FFL
 
 | |
1FWM
 
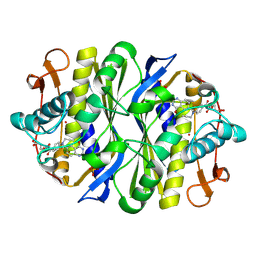 | | Crystal structure of the thymidylate synthase R166Q mutant | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Sotelo-Mundo, R.R, Changchien, L, Maley, F, Montfort, W.R. | | Deposit date: | 2000-09-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thymidylate synthase mutant R166Q: Structural basis for the nearly complete loss of catalytic activity.
J.Biochem.Mol.Toxicol., 20, 2006
|
|
6UKA
 
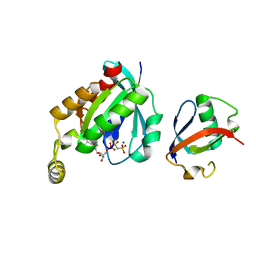 | | Crystal structure of RHOG and ELMO complex | | Descriptor: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jo, C.H, Killoran, R.C, Smith, M.J. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
7MTB
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | Descriptor: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
8E2M
 
 | | Bruton's tyrosine kinase (BTK) with compound 13 | | Descriptor: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | Authors: | Alexander, R, Milligan, C.M. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
