5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
3LPK
 
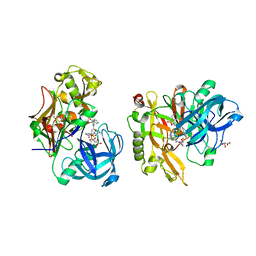 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LNK
 
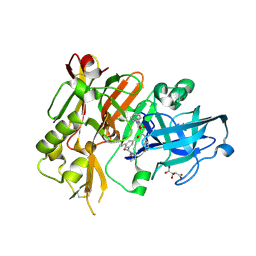 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPJ
 
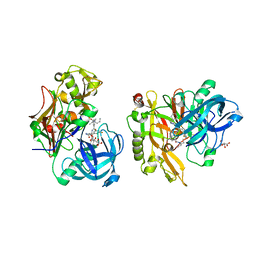 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4ZV8
 
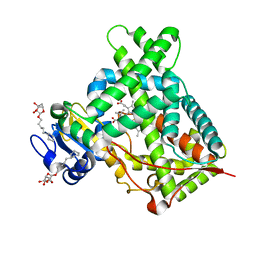 | | Structure of CYP2B6 (Y226H/K262R) with additional mutation Y244W in complex with alpha-Pinene | | Descriptor: | (+)-alpha-Pinene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Liu, J, Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Coumarin Derivatives as Substrate Probes of Mammalian Cytochromes P450 2B4 and 2B6: Assessing the Importance of 7-Alkoxy Chain Length, Halogen Substitution, and Non-Active Site Mutations.
Biochemistry, 55, 2016
|
|
5I9D
 
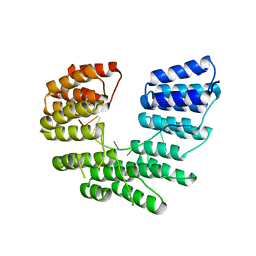 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
7BYF
 
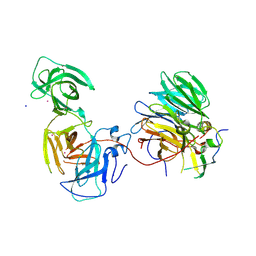 | | The crystal structure of mouse ORF10-Rae1-Nup98 complex | | Descriptor: | 10 protein, MERCURY (II) ION, Peptidase S59 domain-containing protein, ... | | Authors: | Gao, P, Feng, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DDS
 
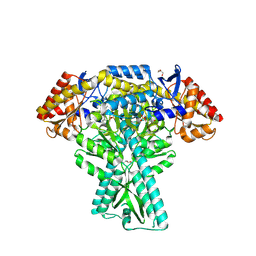 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
8XKF
 
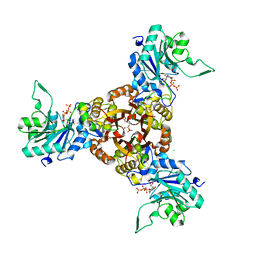 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
6JYZ
 
 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
8XKG
 
 | | Crystal structure of Acinetobacter baumannii IspD | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XHU
 
 | | Crystal structure of Helicobacter pylori IspDF | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
5I9H
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5DDW
 
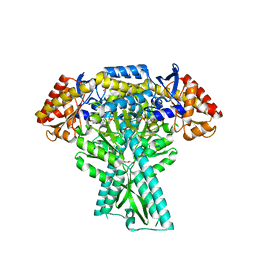 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
7KJS
 
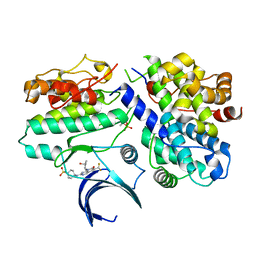 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
5DDU
 
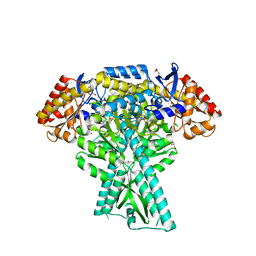 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5I9F
 
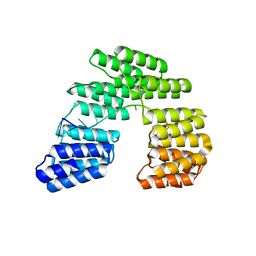 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5BOY
 
 | |
5BPA
 
 | |
5BOT
 
 | |
8JZ4
 
 | | Crystal structure of AetF in complex with FAD and 5-bromo-L-tryptophan | | Descriptor: | 5-bromo-L-tryptophan, AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JU9
 
 | |
8JZ2
 
 | | Crystal structure of AetF in complex with FAD | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ3
 
 | | Crystal structure of AetF in complex with FAD and L-tryptophan | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
