6OED
 
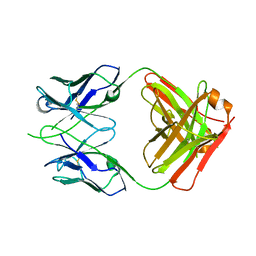 | | CRYSTAL STRUCTURE OF THE RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB | | Descriptor: | CH55 Fab heavy chain, CH55 Fab light chain | | Authors: | Yan, F, Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
7WJR
 
 | |
6C2V
 
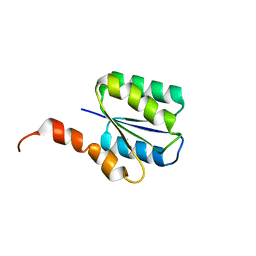 | | Solution structure of a phosphate-loop protein | | Descriptor: | phosphate-loop protein | | Authors: | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | Deposit date: | 2018-01-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C2U
 
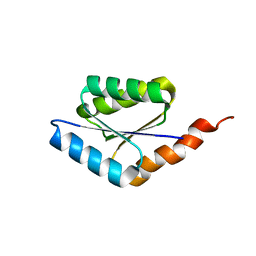 | | Solution structure of a phosphate-loop protein | | Descriptor: | phosphate-loop protein | | Authors: | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | Deposit date: | 2018-01-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1BG8
 
 | |
1C5E
 
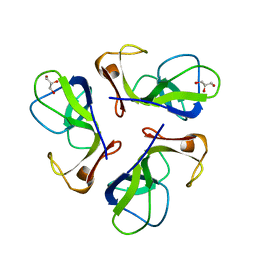 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
2KML
 
 | |
3EZM
 
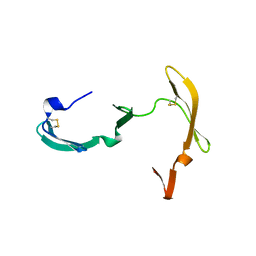 | | CYANOVIRIN-N | | Descriptor: | PROTEIN (CYANOVIRIN-N) | | Authors: | Yang, F, Wlodawer, A. | | Deposit date: | 1998-12-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping.
J.Mol.Biol., 288, 1999
|
|
5T3Y
 
 | |
4Z6G
 
 | | Structure of NT domain | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, PHOSPHATE ION | | Authors: | Yang, F, Zhang, Y. | | Deposit date: | 2015-04-05 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun, 7, 2016
|
|
1SPE
 
 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
3P4L
 
 | |
5GIX
 
 | | Human serum albumin-Palmitic acid-Fe(Hn3piT)Cl2 | | Descriptor: | 14-piperidin-1-yl-11-oxa-13$l^{3}-thia-15,16$l^{4}-diaza-12$l^{3}-ferratetracyclo[8.7.0.0^{2,7}.0^{12,16}]heptadeca-1(10),2(7),3,5,8,13,16-heptaene, PALMITIC ACID, Serum albumin | | Authors: | Yang, F, Qi, J, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Developing Anticancer Ferric Prodrugs Based on the N-Donor Residues of Human Serum Albumin Carrier IIA Subdomain
J. Med. Chem., 59, 2016
|
|
5GIY
 
 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
3UIV
 
 | | Human serum albumin-myristate-amantadine hydrochloride complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Ma, Z, Ma, L, Yang, G. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the drug-binding specificity of human serum albumin
TO BE PUBLISHED
|
|
2NAX
 
 | | Structure of CCHC zinc finger domain of Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3'-end processing.
RNA, 23, 2017
|
|
2I8L
 
 | |
4ZEF
 
 | |
2L2Q
 
 | |
2LBB
 
 | |
2LJU
 
 | |
5YB1
 
 | | Structure and function of human serum albumin-metal agent complex | | Descriptor: | PALMITIC ACID, Serum albumin, ~{N},~{N},12-trimethyl-3$l^{3}-thia-1$l^{4},5,6$l^{4}-triaza-2$l^{3}-cupratricyclo[6.4.0.0^{2,6}]dodeca-1(8),3,6,9,11-pentaen-4-amine | | Authors: | Yang, F, Wang, T, Wang, J, Gou, Y, Zhang, Z.L. | | Deposit date: | 2017-09-02 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developing an Anticancer Copper(II) Multitarget Pro-Drug Based on the His146 Residue in the IB Subdomain of Modified Human Serum Albumin.
Mol. Pharm., 15, 2018
|
|
1UA3
 
 | |
