4N1U
 
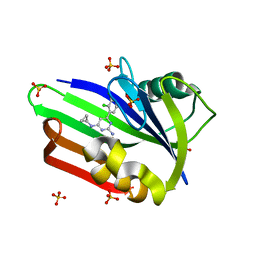 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
7VSQ
 
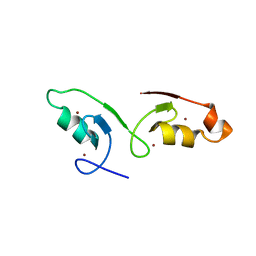 | |
7VSP
 
 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
2R3Q
 
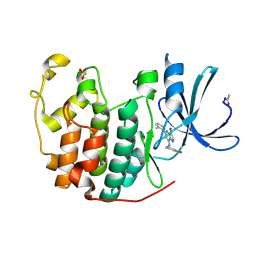 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-((3-bromo-5-o-tolylpyrazolo[1,5-a]pyrimidin-7-ylamino)methyl)pyridine 1-oxide, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
6JOO
 
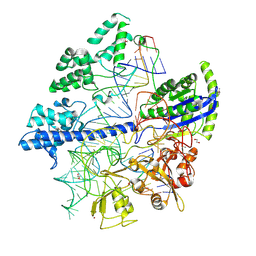 | | Crystal structure of Corynebacterium diphtheriae Cas9 in complex with sgRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein,CRISPR-associated endonuclease Cas9, Guide RNA, ... | | Authors: | Hirano, S, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-03-22 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Nat Commun, 10, 2019
|
|
3CQW
 
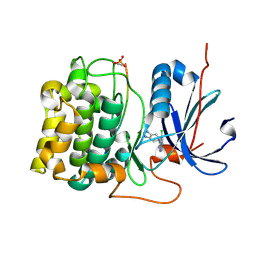 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, Glycogen synthase kinase-3 beta, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5IKI
 
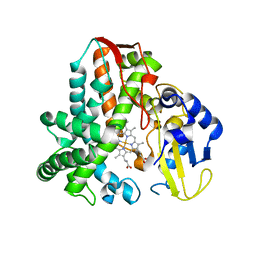 | | CYP106A2 WITH SUBSTRATE ABIETIC ACID | | Descriptor: | Abietic acid, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Janocha, S, Carius, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
3CQU
 
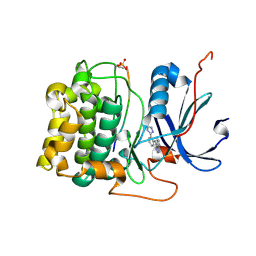 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2FZ4
 
 | |
7E24
 
 | |
7E28
 
 | |
4OO8
 
 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
4KN0
 
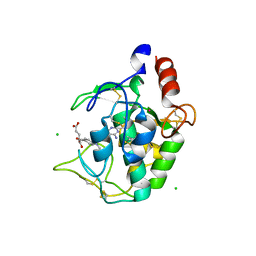 | | Human folate receptor beta (FOLR2) in complex with the antifolate methotrexate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Folate receptor beta, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JVE
 
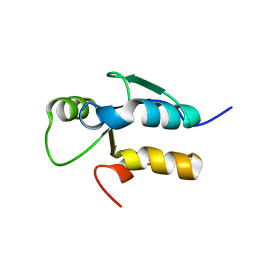 | |
4JPH
 
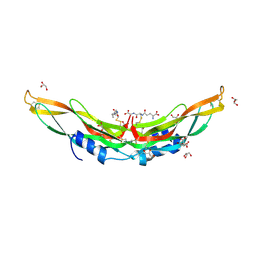 | | Crystal structure of Protein Related to DAN and Cerberus (PRDC) | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Deng, X, Nolan, K.T, Kattamuri, C, Thompson, T.B. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism.
Structure, 21, 2013
|
|
4KMY
 
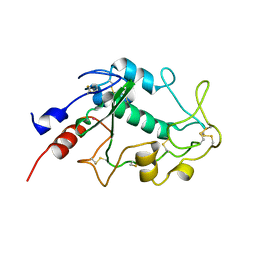 | | Human folate receptor beta (FOLR2) at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor beta, POTASSIUM ION | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KN2
 
 | | Human folate receptor beta (FOLR2) in complex with antifolate pemetrexed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, CHLORIDE ION, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Z72
 
 | |
4KM7
 
 | | Human folate receptor alpha (FOLR1) at acidic pH, triclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor alpha, POTASSIUM ION | | Authors: | Kovach, A.R, Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KM6
 
 | | Human folate receptor alpha (FOLR1) at acidic pH, orthorhombic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Folate receptor alpha | | Authors: | Singh, M, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KN1
 
 | |
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7FJD
 
 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
