6CS4
 
 | |
6CS6
 
 | |
6CS5
 
 | |
1AOE
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | Descriptor: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
6CRP
 
 | |
6CSH
 
 | |
6CRR
 
 | |
6CSA
 
 | |
6CSG
 
 | |
6CS3
 
 | |
6CRS
 
 | |
1CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
5XM9
 
 | |
185D
 
 | |
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
7U1Z
 
 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
5XMA
 
 | |
7UBY
 
 | | Structure of the GTD domain of Clostridium difficile toxin A in complex with VHH AH3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Glucosyltransferase TcdA, ... | | Authors: | Chen, B, Rongsheng, J, Kay, P. | | Deposit date: | 2022-03-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.
Front Immunol, 13, 2022
|
|
1AI9
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
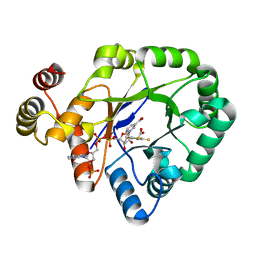 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
6MXR
 
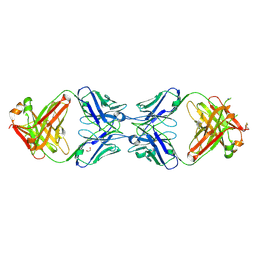 | |
4CJN
 
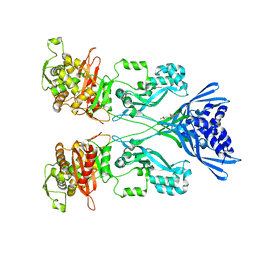 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
5JYK
 
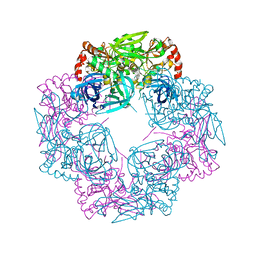 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
