5UKR
 
 | |
5UKO
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522IA Fab | | Descriptor: | DH522IA Fab fragment heavy chain, DH522IA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
4O04
 
 | | Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease | | Descriptor: | 4-(2,7,7-trimethyl-5-oxo-1,2,3,4,5,6,7,8-octahydro-9H-beta-carbolin-9-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
4O0B
 
 | | Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease | | Descriptor: | 8-cyclopentyl-6-(3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-3,4-dihydroisoquinolin-1(2H)-one, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J.T. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
6SZC
 
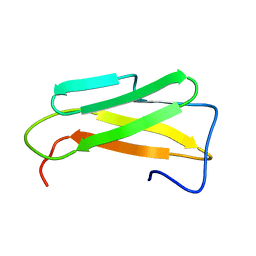 | |
4O05
 
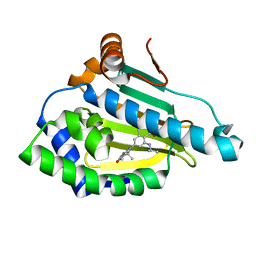 | | Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease | | Descriptor: | 2,7,7-trimethyl-9-[1-oxo-8-(propan-2-ylamino)-1,2,3,4-tetrahydroisoquinolin-6-yl]-1,2,3,4,6,7,8,9-octahydro-5H-beta-carbolin-5-one, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J.T. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
7U0F
 
 | | HIV-1 Rev in complex with tubulin | | Descriptor: | Protein Rev, Tubulin alpha-1A chain, Tubulin beta chain | | Authors: | Eren, E. | | Deposit date: | 2022-02-18 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of microtubule depolymerization by the kinesin-like activity of HIV-1 Rev.
Structure, 31, 2023
|
|
7LPR
 
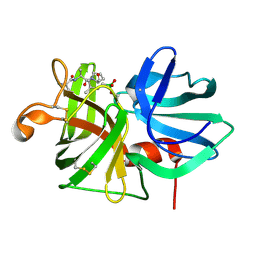 | |
1BMB
 
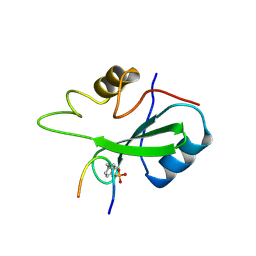 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
6UFY
 
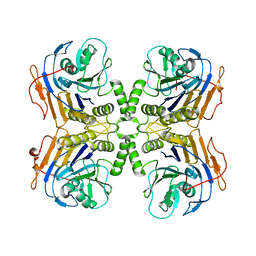 | | B. theta Bile Salt Hydrolase | | Descriptor: | Choloylglycine hydrolase | | Authors: | Seegar, T.C.M. | | Deposit date: | 2019-09-25 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Development of a covalent inhibitor of gut bacterial bile salt hydrolases.
Nat.Chem.Biol., 16, 2020
|
|
1BM2
 
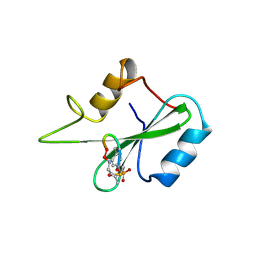 | |
5SWK
 
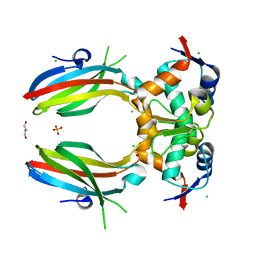 | |
6SSI
 
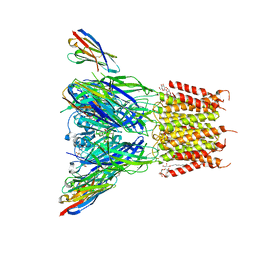 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
6UH4
 
 | | B. theta Bile Salt Hydrolase with covalent inhibitor | | Descriptor: | (5R,6R)-6-[(1S,2R,4aS,4bS,7R,8aS,10R,10aS)-7,10-dihydroxy-1,2,4b-trimethyltetradecahydrophenanthren-2-yl]-5-methylheptan-2-one, Choloylglycine hydrolase | | Authors: | Seegar, T.C.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Development of a covalent inhibitor of gut bacterial bile salt hydrolases.
Nat.Chem.Biol., 16, 2020
|
|
4O3Z
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-95-Ala from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O3Y
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Arg-179-Glu from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O4X
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) double mutant Tyr-167-Ala and Trp-176-Ala from Haemophilus parasuis Hp5 | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O3X
 
 | |
4O09
 
 | | Identification of novel HSP90 / isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington s disease | | Descriptor: | 8-(2-methylpropyl)-6-(3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)-3,4-dihydroisoquinolin-1(2H)-one, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J.T. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
1GBK
 
 | |
1GBB
 
 | |
5DCQ
 
 | | Crystal structure of bacterial adhesin, FNE from Streptococcus equi spp. equi. | | Descriptor: | FORMIC ACID, Fibronectin-binding protein, artificial repeat proteins (alphaREP3) | | Authors: | Tiouajni, M, Graille, M, van Tilbeurgh, H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional analysis of the fibronectin-binding protein FNE from Streptococcus equi spp. equi.
FEBS J., 281, 2014
|
|
4O3W
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-63-Ala from Actinobacillus suis H57 | | Descriptor: | GLYCEROL, SULFATE ION, Transferrin binding protein B | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O07
 
 | | Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease | | Descriptor: | 2,7,7-trimethyl-9-[8-(2-methylpropyl)-1-oxo-1,2,3,4-tetrahydroisoquinolin-6-yl]-1,2,3,4,6,7,8,9-octahydro-5H-beta-carbolin-5-one, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J.T. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
4O4U
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Trp-176-Ala from Haemophilus parasuis Hp5 | | Descriptor: | GLYCEROL, TbpB | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
