6B1B
 
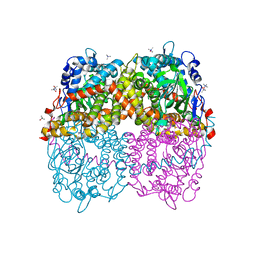 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | Authors: | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | Deposit date: | 2017-09-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
2X2I
 
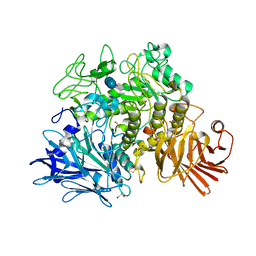 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
5YZ3
 
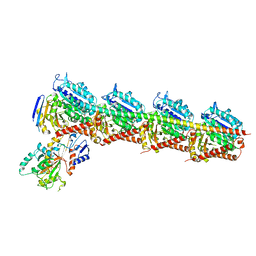 | | Crystal structure of T2R-TTL-28 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-12-12 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | A Novel Microtubule Inhibitor Overcomes Multidrug Resistance in Tumors.
Cancer Res., 78, 2018
|
|
6T2E
 
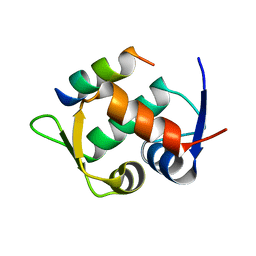 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2D
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2X2H
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2X2J
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
3OCG
 
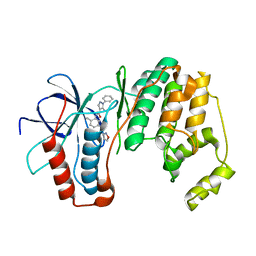 | | P38 Alpha kinase complexed with a 5-amino-pyrazole based inhibitor | | Descriptor: | 5-amino-N-[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]-1-phenyl-1H-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 5-Amino-pyrazoles as potent and selective p38α inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1IDZ
 
 | |
1IDY
 
 | |
8ZBQ
 
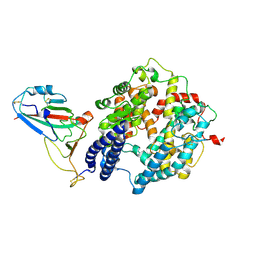 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
4XX4
 
 | |
4XX3
 
 | |
6AFR
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with 5-((4-fluoro-1H-imidazol-1-yl)methyl)quinolin-8-ol | | Descriptor: | 5-[(4-fluoranylimidazol-1-yl)methyl]quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Xing, J, Zhang, R.K, Zheng, M.Y, Luo, C, Jiang, X.R. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Rational design of 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors
Eur J Med Chem, 163, 2018
|
|
4YDV
 
 | | STRUCTURE OF THE ANTIBODY 7B2 THAT CAPTURES HIV-1 VIRIONS | | Descriptor: | HIV ANTIBODY 7B2 HEAVY CHAIN,IgG H chain, HIV ANTIBODY 7B2 LIGHT CHAIN,Ig kappa chain C region, HIV GP41 PEPTIDE GP41(596-606) | | Authors: | Nicely, N.I, Pemble IV, C.W. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-12 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Plos Pathog., 11, 2015
|
|
3L8X
 
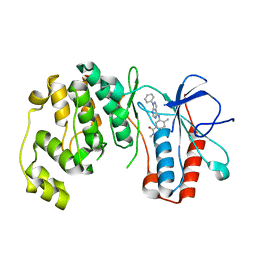 | | P38 alpha kinase complexed with a pyrazolo-pyrimidine based inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N,4-dimethyl-3-[(1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Sack, J.S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pyrazolo-Pyrimidines: A Novel Heterocyclic Scaffold for Potent and Selective P38 Alpha Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1JL4
 
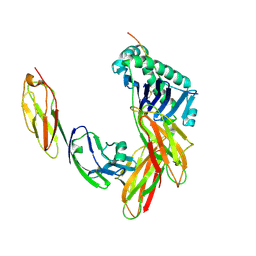 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5IIE
 
 | |
5YPR
 
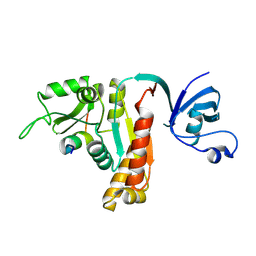 | | Crystal Structure of PSD-95 SH3-GK domain in complex with a synthesized inhibitor | | Descriptor: | Disks large homolog 4, Synthesized GK inhibitor | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhu, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
1L8R
 
 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
5C45
 
 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6M)-2-[(3S)-1-{[2-(methylamino)pyrimidin-5-yl]methyl}piperidin-3-yl]-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Selective small-molecule inhibition of an RNA structural element.
Nature, 526, 2015
|
|
8H08
 
 | |
8H07
 
 | |
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
