4U44
 
 | | MAP4K4 in complex with inhibitor (compound 16) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-phenyl-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U43
 
 | | MAP4K4 in complex with inhibitor (compound 6) | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 4, N-(pyridin-3-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U45
 
 | | MAP4K4 in complex with inhibitor (compound 25) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7DWC
 
 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
6UY0
 
 | |
4J6O
 
 | | Crystal Structure of the Phosphatase Domain of C. thermocellum (Bacterial) PnkP | | Descriptor: | CITRIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wang, L, Smith, P, Shuman, S. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the 2',3' phosphatase component of the bacterial Pnkp-Hen1 RNA repair system.
Nucleic Acids Res., 41, 2013
|
|
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
3DBZ
 
 | | human surfactant protein D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of recombinant surfactant protein D with lipopolysaccharide: conformation and orientation of bound protein by IRRAS and simulations.
Biochemistry, 47, 2008
|
|
7TYJ
 
 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
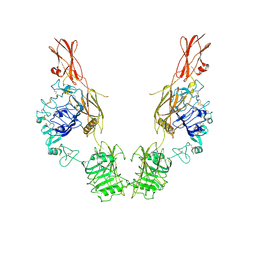 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
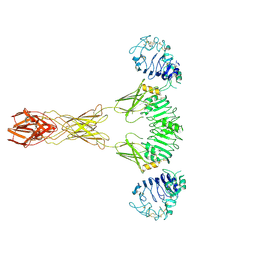 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7LRT
 
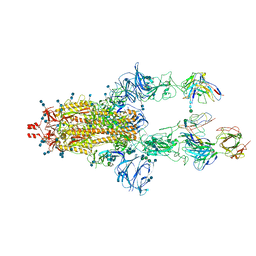 | |
7LRS
 
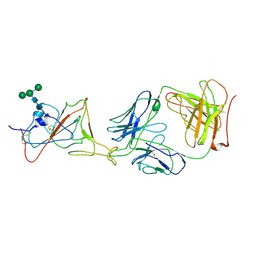 | |
7CBZ
 
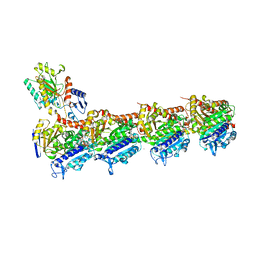 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
6L5T
 
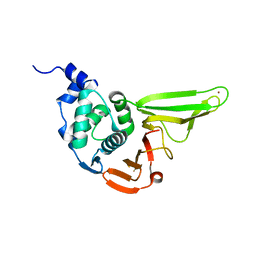 | | The crystal structure of SADS-CoV Papain Like protease | | Descriptor: | Peptidase C16, ZINC ION | | Authors: | Fan, C.P. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical characterization of SADS-CoV papain-like protease 2.
Protein Sci., 29, 2020
|
|
7L8O
 
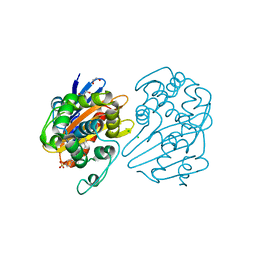 | | OXA-48 bound by Compound 4.3 | | Descriptor: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
4ANW
 
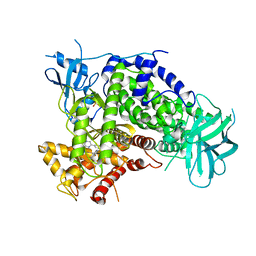 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-6-{4-CHLORO-3-[(2,3-DIFLUOROPHENYL)SULFAMOYL]PHENYL}-N-METHYLPYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
7LF6
 
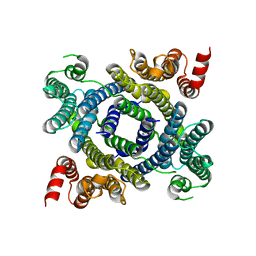 | | Structure of lysosomal membrane protein | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Shen, C, Fu, T.M, Wang, L.F, Rawson, S, Wu, H. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | pH regulates potassium conductance and drives a constitutive proton current in human TMEM175.
Sci Adv, 8, 2022
|
|
6E99
 
 | |
6E9W
 
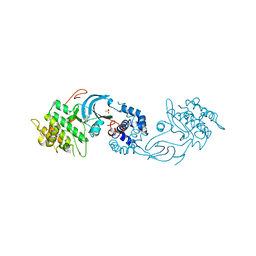 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6E9L
 
 | |
6ED6
 
 | |
