7ZJY
 
 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZK0
 
 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
2W4L
 
 | | Human dCMP deaminase | | Descriptor: | CHLORIDE ION, DEOXYCYTIDYLATE DEAMINASE, ZINC ION | | Authors: | Siponen, M.I, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schuler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Nordlund, P. | | Deposit date: | 2008-11-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Human Dcmp Deaminase
To be Published
|
|
8QNH
 
 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
7TPR
 
 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
6QYI
 
 | | Structure of HPAB from E.coli in complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
5LKB
 
 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
8QN5
 
 | | M. tuberculosis salicylate synthase MbtI in complex with methyl-AMT (new crystal form) | | Descriptor: | 3-{[(1Z)-1-carboxyprop-1-en-1-yl]oxy}-2-hydroxybenzoic acid, AMMONIUM ION, CITRATE ANION, ... | | Authors: | Mori, M, Villa, S, Meneghetti, M, Bellinzoni, M. | | Deposit date: | 2023-09-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Structural Study of a New MbtI-Inhibitor Complex: Towards an Optimized Model for Structure-Based Drug Discovery.
Pharmaceuticals, 16, 2023
|
|
5LPI
 
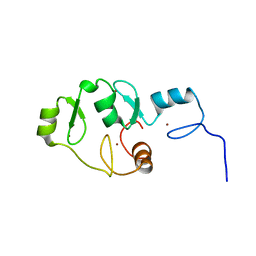 | |
7TRJ
 
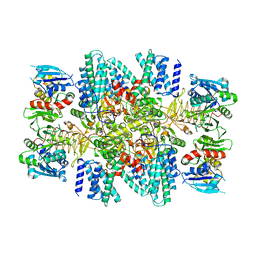 | | The eukaryotic translation initiation factor 2B from Homo sapiens with a H160D mutation in the beta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Schoof, M, Lawrence, R, Boone, M, Frost, A, Walter, P. | | Deposit date: | 2022-01-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A point mutation in the nucleotide exchange factor eIF2B constitutively activates the integrated stress response by allosteric modulation.
Elife, 11, 2022
|
|
5LTO
 
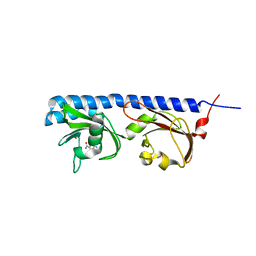 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5M3L
 
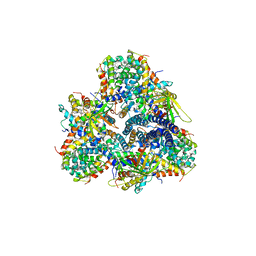 | | Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris hemoglobin | | Descriptor: | Extracellular globin-2, Extracellular globin-3, Extracellular globin-4, ... | | Authors: | Afanasyev, P, Linnemayr-Seer, C, Ravelli, R.B.G, Matadeen, R, De Carlo, S, Alewijnse, B, Portugal, R.V, Pannu, N.S, Schatz, M, van Heel, M. | | Deposit date: | 2016-10-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris haemoglobin.
IUCrJ, 4, 2017
|
|
6GAA
 
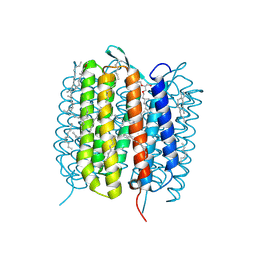 | | BACTERIORHODOPSIN, 430 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6ZBA
 
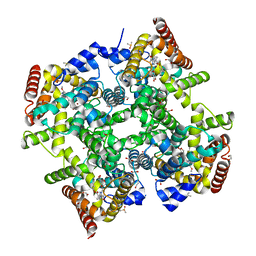 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
6GAE
 
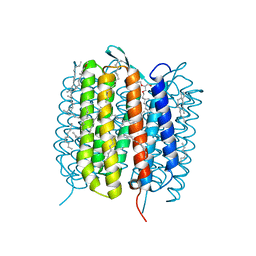 | | BACTERIORHODOPSIN, 560 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
7TJ0
 
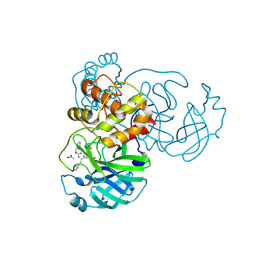 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
5GJD
 
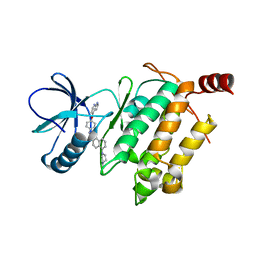 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 2 | | Descriptor: | 1-(4-((1H-pyrrolo[2,3-b]pyridin-4-yl)oxy)phenyl)-3-(5-(4-methylpiperazin-1-yl)naphthalen-2-yl)urea, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
8QTG
 
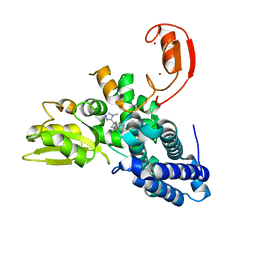 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTH
 
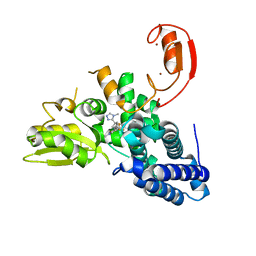 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTJ
 
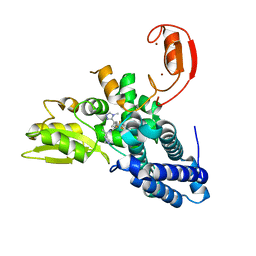 | | Crystal structure of Cbl-b in complex with an allosteric inhibitor (compound 30) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1~{R})-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QTK
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 31) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1S)-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
6R8G
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
6AGT
 
 | | Crystal structure of PfKRS complexed with chromone inhibitor | | Descriptor: | COBALT (II) ION, FORMIC ACID, LYSINE, ... | | Authors: | Yogavel, M, Sharma, A, Sharma, A, Baragana, B, Walpole, C. | | Deposit date: | 2018-08-14 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
