7CV7
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSC
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | Descriptor: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5TUY
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor MS0124 | | Descriptor: | 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2016-11-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
7FDO
 
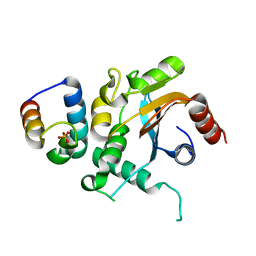 | |
5EGS
 
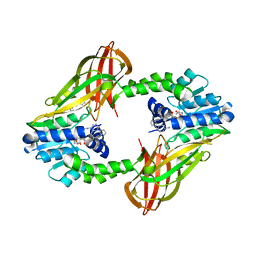 | | Human PRMT6 with bound fragment-type inhibitor | | Descriptor: | 2-[4-(phenylmethyl)piperidin-1-yl]ethanamine, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Steuber, H, Egner, U, Kania, J, Wu, H, Brown, P.J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Potent Class I Protein Arginine Methyltransferase Fragment Inhibitor.
J.Med.Chem., 59, 2016
|
|
4FU6
 
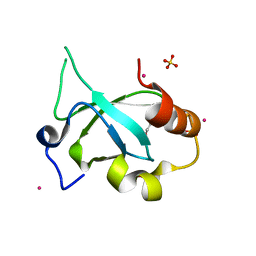 | | Crystal structure of the PSIP1 PWWP domain | | Descriptor: | GLYCEROL, PC4 and SFRS1-interacting protein, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Xu, C, Wu, H, Dong, A, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the nucleosome-binding PWWP domain.
Trends Biochem.Sci., 39, 2014
|
|
6E16
 
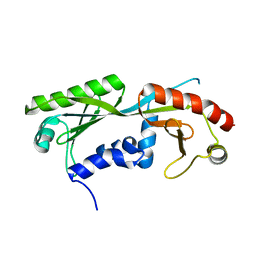 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6E24
 
 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6CD7
 
 | | Crystal structure of APH(2")-IVa in complex with plazomicin | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, APH(2'')-Id, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Di Leo, R, Savchenko, A, Satchell, K.J, Joachimiak, J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5WCJ
 
 | | Crystal Structure of Human Methyltransferase-like protein 13 in complex with SAH | | Descriptor: | Methyltransferase-like protein 13, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Loppnau, P, Seitova, A, Hutchinson, A, Hunt, B, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates.
Nat Commun, 9, 2018
|
|
5C7J
 
 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
4MYO
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
5CYV
 
 | | Crystal structure of CouR from Rhodococcus jostii RHA1 bound to p-coumaroyl-CoA | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The activity of CouR, a MarR family transcriptional regulator, is modulated through a novel molecular mechanism.
Nucleic Acids Res., 44, 2016
|
|
4PZI
 
 | | Zinc finger region of MLL2 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Histone-lysine N-methyltransferase 2B, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Liu, K, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4O62
 
 | | CW-type zinc finger of ZCWPW2 in complex with the amino terminus of histone H3 | | Descriptor: | Histone H3.3, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
4O61
 
 | | Structure of human ALKBH5 crystallized in the presence of citrate | | Descriptor: | CITRIC ACID, GLYCEROL, RNA demethylase ALKBH5, ... | | Authors: | Tempel, W, Chao, X, Liu, K, Dong, A, Cerovina, T, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation.
J.Biol.Chem., 289, 2014
|
|
4OCT
 
 | | Crystal structure of human ALKBH5 crystallized in the presence of Mn^{2+} and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA demethylase ALKBH5, ... | | Authors: | Tempel, W, Chao, X, Liu, K, Dong, A, Cerovina, T, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-09 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation.
J.Biol.Chem., 289, 2014
|
|
7D97
 
 | |
7D95
 
 | |
7D40
 
 | |
7D96
 
 | |
6WY0
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WXZ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6WYD
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-12 (AKA; 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyrid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-benzyl-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
