6U09
 
 | |
6U98
 
 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 6 | | Descriptor: | 3-{[(3R)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha, POTASSIUM ION, ... | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U9B
 
 | | Hsp90a NTD covalently bound to sulfonyl fluoride 5 at K58 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U06
 
 | |
6U99
 
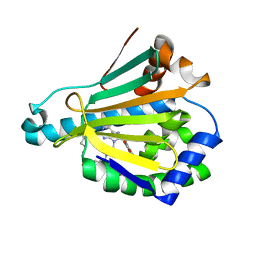 | | Hsp90a NTD covalently bound to sulfonyl fluoride probe 1 at K58 | | Descriptor: | 3-{[(3-{6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}propyl)amino]methyl}benzene-1-sulfinic acid, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U9A
 
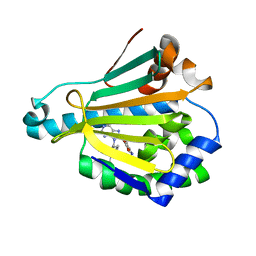 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 5 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
8SCB
 
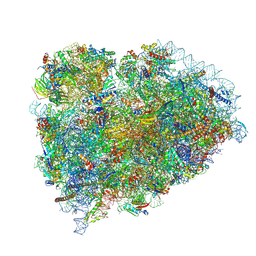 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
5U8L
 
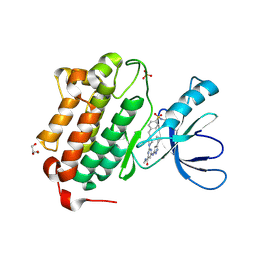 | | Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride probe XO44 | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
7ZL3
 
 | |
3QBV
 
 | | Structure of designed orthogonal interaction between CDC42 and nucleotide exchange domains of intersectin | | Descriptor: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, Intersectin-1 | | Authors: | Kapp, G.T, Remenyi, A, Lim, W.A, Kortemme, T. | | Deposit date: | 2011-01-14 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LV2
 
 | |
4LV1
 
 | |
4LV3
 
 | | AmpC beta-lactamase in complex with (3,5-di-tert-butylphenyl) boronic acid | | Descriptor: | (3,5-di-tert-butylphenyl)boronic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | London, N, Eidam, O, Shoichet, B.K. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Covalent docking of large libraries for the discovery of chemical probes.
Nat.Chem.Biol., 10, 2014
|
|
8DIC
 
 | |
8DIB
 
 | |
8DIG
 
 | |
8DII
 
 | |
8DIE
 
 | |
8DIF
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DID
 
 | |
4LV0
 
 | |
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
