2LZE
 
 | | Ligase 10C | | Descriptor: | ZINC ION, a primordial catalytic fold generated by in vitro evolution | | Authors: | Chao, F, Morelli, A, Haugner, J, Churchfield, L, Hagmann, L, Shi, L, Masterson, L, Sarangi, R, Veglia, G, Seelig, B. | | Deposit date: | 2012-10-01 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of a primordial catalytic fold generated by in vitro evolution.
Nat.Chem.Biol., 9, 2013
|
|
2M3G
 
 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy | | Descriptor: | Anabaena Sensory Rhodopsin, RETINAL | | Authors: | Wang, S, Munro, R.A, Shi, L, Kawamura, I, Okitsu, T, Wada, A, Kim, S, Jung, K, Brown, L.S, Ladizhansky, V. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-16 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein.
Nat.Methods, 10, 2013
|
|
2MT6
 
 | | Solution structure of the human ubiquitin conjugating enzyme Ube2w | | Descriptor: | Ubiquitin-conjugating enzyme E2 W | | Authors: | Vittal, V, Shi, L, Wenzel, D.M, Brzovic, P.S, Klevit, R.E. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Nat.Chem.Biol., 11, 2015
|
|
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
2ONC
 
 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
2KYV
 
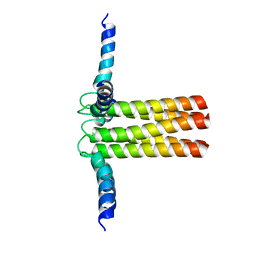 | | Hybrid solution and solid-state NMR structure ensemble of phospholamban pentamer | | Descriptor: | Phospholamban | | Authors: | Verardi, R, Shi, L, Traaseth, N.J, Veglia, G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LPF
 
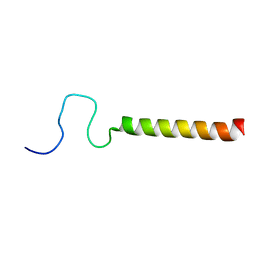 | | R state structure of monomeric phospholamban (C36A, C41F, C46A) | | Descriptor: | Cardiac phospholamban | | Authors: | De Simone, A, Montalvao, R.W, Gustavsson, M, Shi, L, Veglia, G, Vendruscolo, M. | | Deposit date: | 2012-02-11 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Excited States of Phospholamban and Shifts in Their Populations upon Phosphorylation.
Biochemistry, 52, 2013
|
|
2M89
 
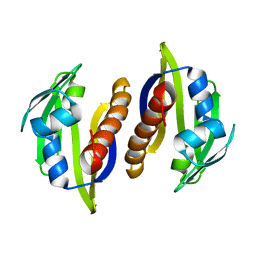 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2N0K
 
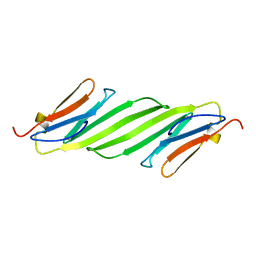 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
2MUU
 
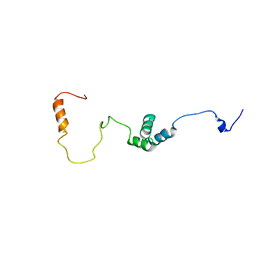 | |
1QYI
 
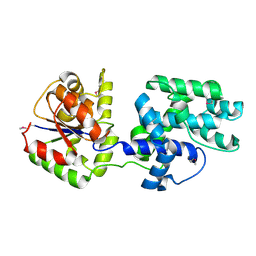 | | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25. | | Descriptor: | hypothetical protein | | Authors: | Kuzin, A.P, Edstrom, W, Ma, L.C, Shin, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-10 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25.
To be Published
|
|
6EKN
 
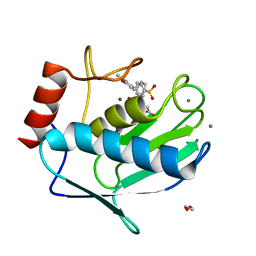 | | Crystal structure of MMP12 in complex with inhibitor BE7. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6ELA
 
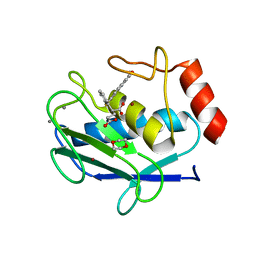 | | Crystal structure of MMP12 in complex with inhibitor BE4. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfanylphenyl]pentanedioic acid, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-09-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.485 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6ESM
 
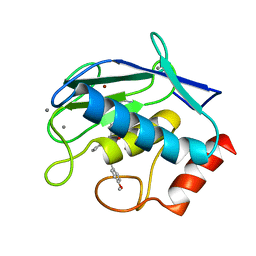 | | Crystal structure of MMP9 in complex with inhibitor BE4. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfanylphenyl]pentanedioic acid, CALCIUM ION, Matrix metalloproteinase-9,Matrix metalloproteinase-9, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-10-23 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
6P4Y
 
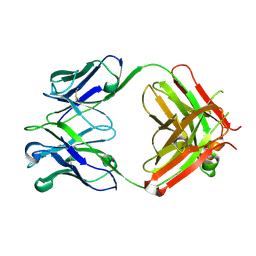 | |
6P67
 
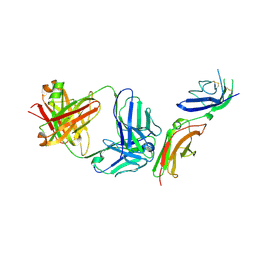 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha 2B8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-IL-7R 2B8 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-06-03 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
6P50
 
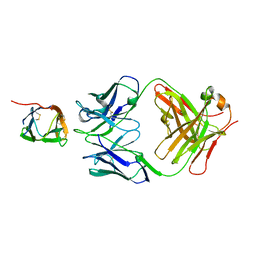 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha Fab 4A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-7 receptor subunit alpha, anti-IL-7R 4A10 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
5MG9
 
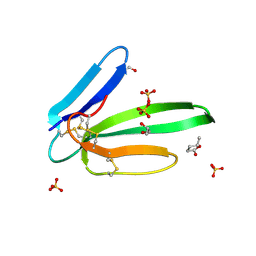 | | Putative Ancestral Mamba toxin 1 (AncTx1-W28R/I38S) | | Descriptor: | 1,2-ETHANEDIOL, AncTx1-W28R/I38S, S-1,2-PROPANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Blanchet, G, Mourier, G, Servent, D. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins.
Sci Rep, 7, 2017
|
|
5DO6
 
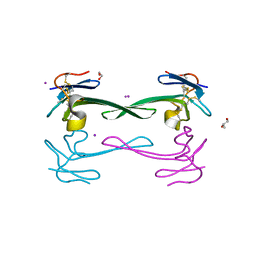 | | Crystal structure of Dendroaspis polylepis venom mambalgin-1 T23A mutant | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Mambalgin-1, ... | | Authors: | Stura, E.A, Tepshi, L, Kessler, P, Gilles, M, Servent, D. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
8SFY
 
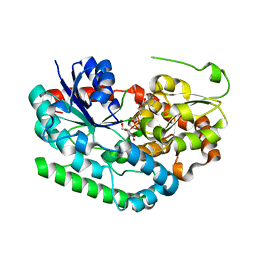 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with UDP-glucose | | Descriptor: | UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Arriaza, R.H, Daneshian, L, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with UDP-glucose
To Be Published
|
|
7S2H
 
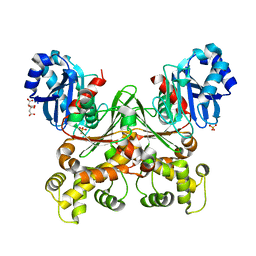 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
7S2P
 
 | | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, SULFATE ION | | Authors: | Carey, S.M, Nettles, R.B, Daneshian, L, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN
To Be Published
|
|
7S2N
 
 | |
7C0M
 
 | | Human cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Zierhut, C, Takizawa, Y, Kim, R, Negishi, L, Uruma, N, Hirai, S, Funabiki, H, Kurumizaka, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the inhibition of cGAS by nucleosomes.
Science, 370, 2020
|
|
