4I6L
 
 | |
4JVG
 
 | | B-Raf Kinase in Complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Thevakumaran, N, Gavory, G, Li, J, Padeganeh, A, Guiral, S, Duchaine, J, Mao, D.Y.L, Bouvier, M, Sicheri, F, Therrien, M. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibitors that stabilize a closed RAF kinase domain conformation induce dimerization.
Nat.Chem.Biol., 9, 2013
|
|
6DJ9
 
 | | Structure of the USP15 DUSP domain in complex with a high-affinity Ubiquitin Variant (UbV) | | Descriptor: | Ubiquitin Variant UbV 15.D, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-05-24 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6DRM
 
 | | OTU domain of Fam105A | | Descriptor: | Inactive ubiquitin thioesterase FAM105A | | Authors: | Ceccarelli, D.F, Sicheri, F, Cordes, S. | | Deposit date: | 2018-06-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | FAM105A/OTULINL Is a Pseudodeubiquitinase of the OTU-Class that Localizes to the ER Membrane.
Structure, 27, 2019
|
|
4DDI
 
 | | Crystal structure of human OTUB1/UbcH5b~Ub/Ub | | Descriptor: | Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D2, Ubiquitin thioesterase OTUB1 | | Authors: | Juang, Y.C, Sanches, M, Sicheri, F. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function.
Mol.Cell, 45, 2012
|
|
4DDG
 
 | | Crystal structure of human OTUB1/UbcH5b~Ub/Ub | | Descriptor: | Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D2, Ubiquitin thioesterase OTUB1 | | Authors: | Juang, Y.C, Sanches, M, Sicheri, F. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2987 Å) | | Cite: | OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function.
Mol.Cell, 45, 2012
|
|
8BW9
 
 | |
8BW8
 
 | |
8D6E
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with RP-6306 | | Descriptor: | (1P)-2-amino-1-(3-hydroxy-2,6-dimethylphenyl)-5,6-dimethyl-1H-pyrrolo[2,3-b]pyridine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6D
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6F
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with Eph receptor inhibitor / compound 41 | | Descriptor: | (1M)-2-amino-1-(5-hydroxy-2-methylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, SULFATE ION | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6C
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 28 | | Descriptor: | (1P)-2-amino-6-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
5IBK
 
 | | Skp1-F-box in complex with a ubiquitin variant | | Descriptor: | F-box/WD repeat-containing protein 7, Polyubiquitin-B, S-phase kinase-associated protein 1,S-phase kinase-associated protein 1 | | Authors: | Orlicky, S, Sicheri, F. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J26
 
 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
5JMV
 
 | | Crystal structure of mjKae1-pfuPcc1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, ... | | Authors: | Wan, L, Sicheri, F. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3864696 Å) | | Cite: | Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis.
Nucleic Acids Res., 44, 2016
|
|
3BS7
 
 | |
3BS5
 
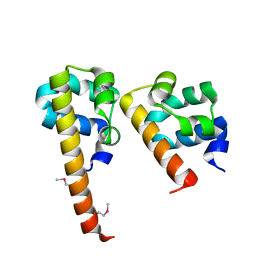 | | Crystal Structure of hCNK2-SAM/dHYP-SAM Complex | | Descriptor: | Connector enhancer of kinase suppressor of ras 2, Protein aveugle | | Authors: | Rajakulendran, T, Ceccarelli, D.F, Kurinov, I, Sicheri, F. | | Deposit date: | 2007-12-22 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3BQ3
 
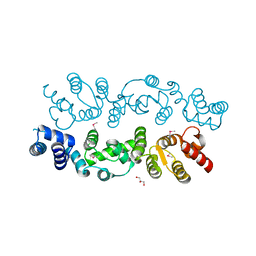 | | Crystal structure of S. cerevisiae Dcn1 | | Descriptor: | Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Chou, Y.C, Sicheri, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dcn1 Functions as a Scaffold-Type E3 Ligase for Cullin Neddylation.
Mol.Cell, 29, 2008
|
|
6ML1
 
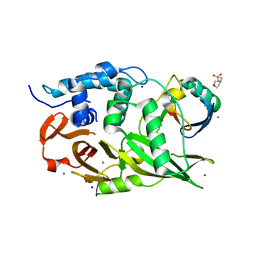 | | Structure of the USP15 deubiquitinase domain in complex with an affinity-matured inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6NJG
 
 | |
6NTD
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 12 of CRAF Kinase protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
6NSQ
 
 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
6NTC
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 1 of CRAF Kinase protein | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
2F8K
 
 | | Sequence specific recognition of RNA hairpins by the SAM domain of Vts1 | | Descriptor: | 5'-R(*UP*AP*AP*UP*CP*UP*UP*UP*GP*AP*CP*AP*GP*AP*UP*U)-3', Protein VTS1 | | Authors: | Aviv, T, Lin, Z, Ben-Ari, G, Smibert, C.A, Sicheri, F. | | Deposit date: | 2005-12-02 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1OPS
 
 | | ICE-BINDING SURFACE ON A TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT | | Descriptor: | TYPE III ANTIFREEZE PROTEIN | | Authors: | Yang, D.S.C, Hon, W.-C, Bubanko, S, Xue, Y, Seetharaman, J, Hew, C.L, Sicheri, F. | | Deposit date: | 1997-11-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of the ice-binding surface on a type III antifreeze protein with a "flatness function" algorithm.
Biophys.J., 74, 1998
|
|
