6E9L
 
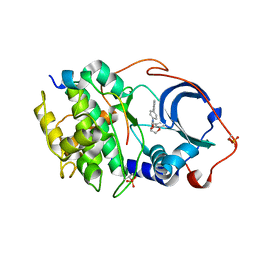 | |
6ED6
 
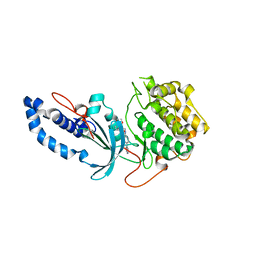 | |
3M6F
 
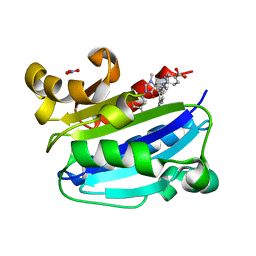 | | CD11A I-domain complexed with 6-((5S,9R)-9-(4-CYANOPHENYL)-3-(3,5-DICHLOROPHENYL)-1-METHYL-2,4-DIOXO-1,3,7- TRIAZASPIRO[4.4]NON-7-YL)NICOTINIC ACID | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, NITRATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small molecule antagonist of leukocyte function associated antigen-1 (LFA-1): structure-activity relationships leading to the identification of 6-((5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]nonan-7-yl)nicotinic acid (BMS-688521).
J.Med.Chem., 53, 2010
|
|
6XDG
 
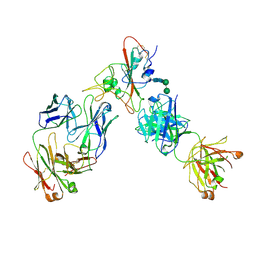 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
6WJL
 
 | | Crystal structure of Glypican-2 core protein in complex with D3 Fab | | Descriptor: | D3 Fab Heavy chain, D3 Fab Light Chain, Glypican-2, ... | | Authors: | Raman, S, Maris, J.M, Bosse, K.R, Julien, J.P. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A GPC2 antibody-drug conjugate is efficacious against neuroblastoma and small-cell lung cancer via binding a conformational epitope.
Cell Rep Med, 2, 2021
|
|
8J2W
 
 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2Y
 
 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2X
 
 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
3I81
 
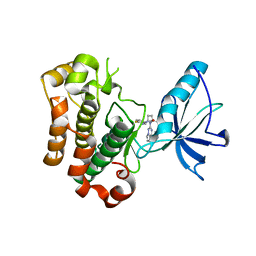 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with BMS-754807 [1-(4-((5-cyclopropyl-1H-pyrazol-3-yl)amino)pyrrolo[2,1-f][1,2,4]triazin-2-yl)-N-(6-fluoro-3-pyridinyl)-2-methyl-L-prolinamide] | | Descriptor: | 1-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}-N-(6-fluoropyridin-3-yl)-2-methyl-L-proli namide, Insulin-like growth factor 1 receptor | | Authors: | Sack, J.S. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development.
J.Med.Chem., 52, 2009
|
|
6E99
 
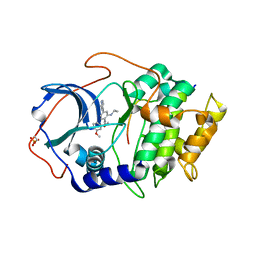 | |
6E9W
 
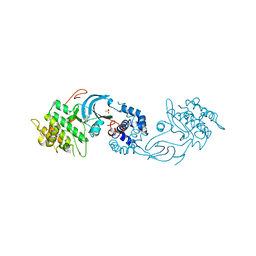 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
8CV5
 
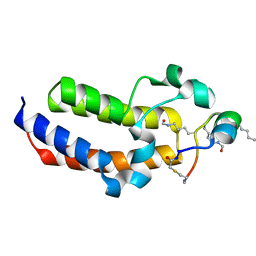 | | Peptide 4.2B in complex with BRD3.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Bromodomain-containing protein 3, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV7
 
 | | Peptide 2.2E in complex with BRD2-BD2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Isoform 3 of Bromodomain-containing protein 2, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV4
 
 | | Peptide 4.2C in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV6
 
 | | Peptide 4.2B in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
7M4S
 
 | |
6WSH
 
 | |
6WW6
 
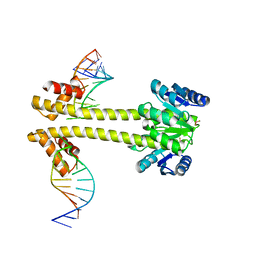 | | Crystal structure of EutV bound to RNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Response regulator, eutP P2 | | Authors: | Ataide, S.F, Walshe, J.L. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural characterization of the ANTAR antiterminator domain bound to RNA.
Nucleic Acids Res., 50, 2022
|
|
8TOQ
 
 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOS
 
 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOR
 
 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOT
 
 | | ACE2-peptide2 complex crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOU
 
 | | ACE2-peptide 2 complex crystal form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Franck, C, Payne, R.J, Christie, M. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
4RNA
 
 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
7L4Z
 
 | | Structure of SARS-CoV-2 spike RBD in complex with cyclic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2, Spike protein S1 | | Authors: | Christie, M, Mackay, J.P, Passioura, T, Payne, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display.
Acs Cent.Sci., 7, 2021
|
|
