5JAF
 
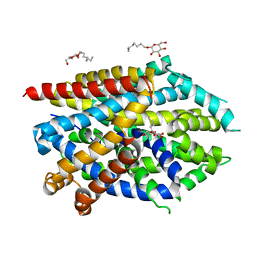 | | LeuT Na+-free Return State, C2 form at pH 5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.021 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAG
 
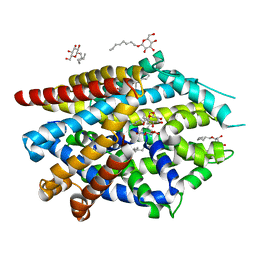 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
6HEF
 
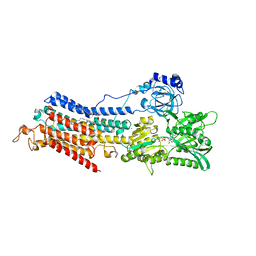 | | Room temperature structure of the (SR)Ca2+-ATPase Ca2-E1-CaAMPPCP form | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Hjorth-Jensen, S, Sorensen, T.L.M, Oksanen, E, Andersen, J.L, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | Membrane-protein crystals for neutron diffraction.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5A3Q
 
 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-AMPPCP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
5A3S
 
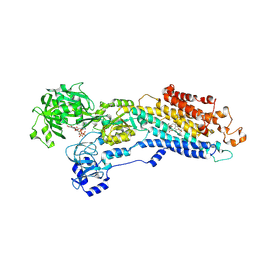 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-ATP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
5A3R
 
 | | Crystal structure of the (SR) Calcium ATPase E2.BeF3- complex bound to TNP-AMPPCP | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SARCOPLASMIC/ENDOPLASMIC RETICULUM CALCIUM ATPASE 1, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
6ROI
 
 | | Cryo-EM structure of the partially activated Drs2p-Cdc50p | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
6ROJ
 
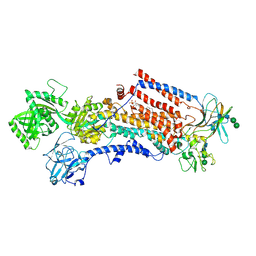 | | Cryo-EM structure of the activated Drs2p-Cdc50p | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
6ROH
 
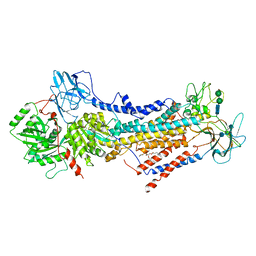 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
4US3
 
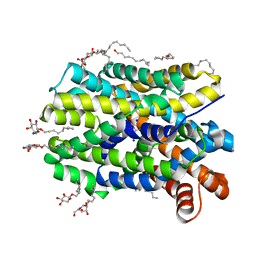 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UU0
 
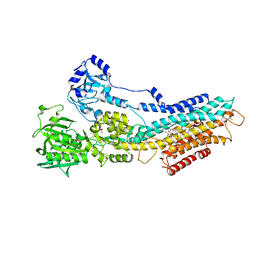 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF 14:1 PC | | Descriptor: | GLYCEROL, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
4US4
 
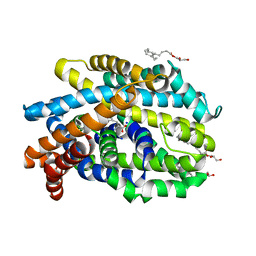 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4US7
 
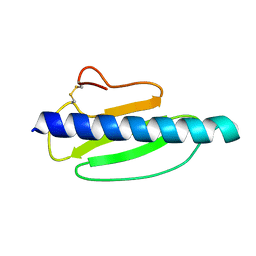 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | Descriptor: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | Authors: | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | Deposit date: | 2014-07-03 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
5O1S
 
 | |
4BEV
 
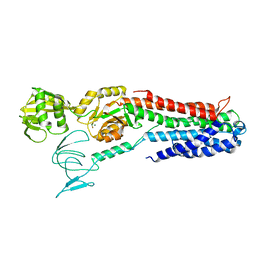 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
4BEW
 
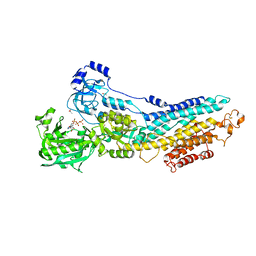 | | SERCA bound to phosphate analogue | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Drachmann, N.D, Mattle, D, Laursen, M, Bublitz, M, Olesen, C, Moeller, J.V, Nissen, P, Morth, J.P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serca Bound to Phosphate Analogue
To be Published
|
|
3N8G
 
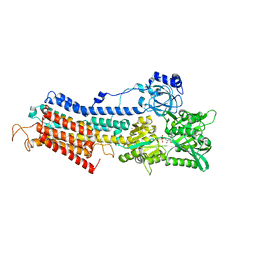 | | Structure of the (SR)Ca2+-ATPase Ca2-E1-CaAMPPCP form | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Olesen, C, Poulsen, H, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 288, 2013
|
|
1K8A
 
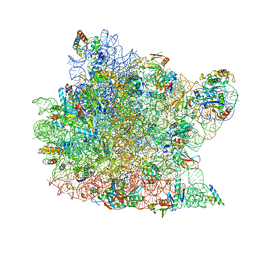 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
4BBJ
 
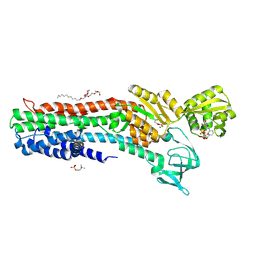 | | Copper-transporting PIB-ATPase in complex with beryllium fluoride representing the E2P state | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mattle, D, Gourdon, P, Nissen, P. | | Deposit date: | 2012-09-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Copper-Transporting P-Type Atpases Use a Unique Ion-Release Pathway
Nat.Struct.Mol.Biol., 21, 2014
|
|
4BYG
 
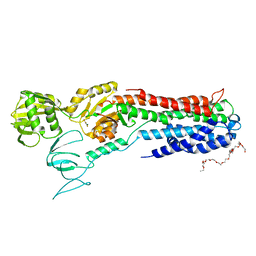 | | ATPase crystal structure | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Pedersen, B.P, Morth, J.P, Wang, J, Gourdon, P, Nissen, P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dephosphorylation of Pib-Type Cu(I)-Atpases as Studied by Metallofluoride Complexes
To be Published
|
|
3F6K
 
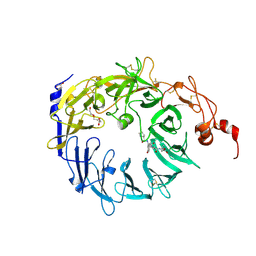 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with neurotensin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neurotensin, ... | | Authors: | Quistgaard, E.M, Madsen, P, Groftehauge, M.K, Nissen, P, Petersen, C.M, Thirup, S. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6ZBV
 
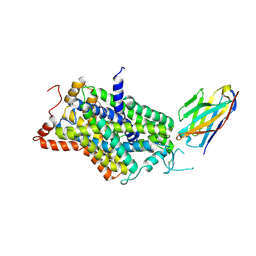 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | Descriptor: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
2P8Z
 
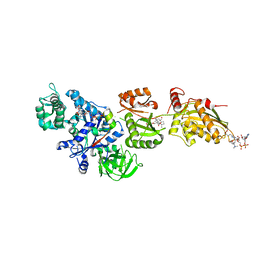 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
6ZPL
 
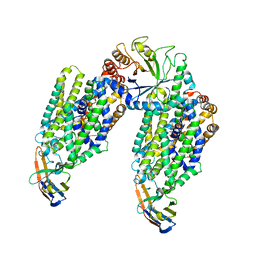 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
2P8W
 
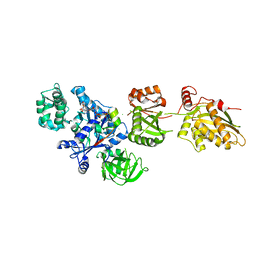 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
