1G1J
 
 | |
1G1I
 
 | |
4NCC
 
 | | Neutralizing antibody to murine norovirus | | Descriptor: | Fab fragment heavy, Fab fragment light | | Authors: | Smith, T, Li, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-19 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization.
J.Virol., 88, 2014
|
|
6M14
 
 | | Crystal Structure of the BARD1 BRCT Mutant | | Descriptor: | BRCA1-associated RING domain protein 1, SULFATE ION | | Authors: | Chen, T, Huang, W.T. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88001835 Å) | | Cite: | BARD1 is an ATPase activating protein for OLA1.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
8IBH
 
 | | Cep57 C-terminal domain | | Descriptor: | Centrosomal protein of 57 kDa | | Authors: | Chen, T, Yeh, H.-W, Cheng, H.-C. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cep57 regulates human centrosomes through multivalent interactions.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WIH
 
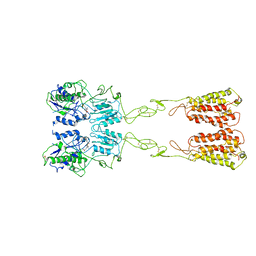 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
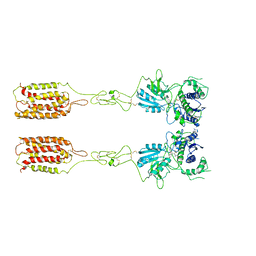 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
3ASW
 
 | |
3AT0
 
 | |
7BZ2
 
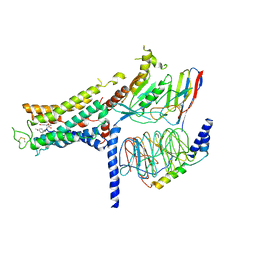 | | Cryo-EM structure of the formoterol-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y.N, Yang, F, Ling, S.L, Lv, P, Zhou, Y.X, Fang, W, Sun, W, Shi, P, Tian, C.L. | | Deposit date: | 2020-04-26 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Single-particle cryo-EM structural studies of the beta2AR-Gs complex bound with a full agonist formoterol.
Cell Discov, 6, 2020
|
|
6P72
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein global domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-04 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7S
 
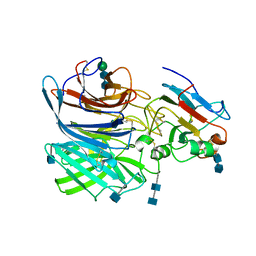 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7Y
 
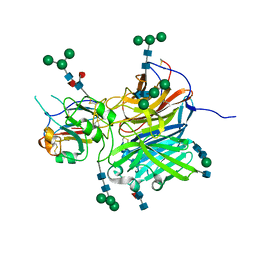 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8FAH
 
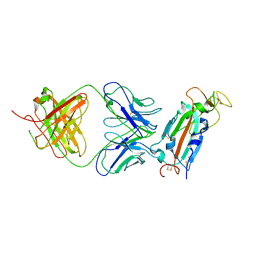 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with SARS-CoV-2 reactive human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.22 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
4YDV
 
 | | STRUCTURE OF THE ANTIBODY 7B2 THAT CAPTURES HIV-1 VIRIONS | | Descriptor: | HIV ANTIBODY 7B2 HEAVY CHAIN,IgG H chain, HIV ANTIBODY 7B2 LIGHT CHAIN,Ig kappa chain C region, HIV GP41 PEPTIDE GP41(596-606) | | Authors: | Nicely, N.I, Pemble IV, C.W. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-12 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Plos Pathog., 11, 2015
|
|
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7X1U
 
 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with an Endogenous Peptide Agonist TRH. | | Descriptor: | Endogenous Peptide Agonist TRH, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
7XAT
 
 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAV
 
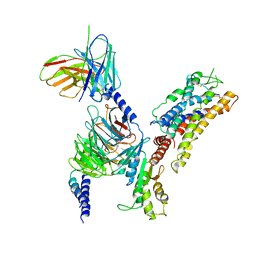 | | Structure of somatostatin receptor 2 bound with lanreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
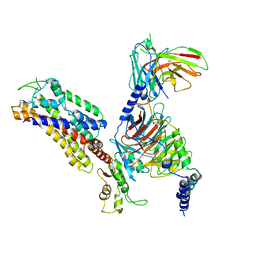 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7X1T
 
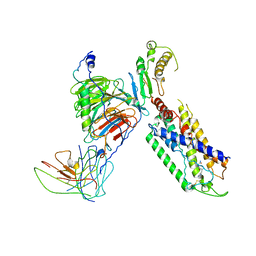 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with Taltirelin. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ScFv16, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
3NYB
 
 | |
7XWO
 
 | | Neurokinin A bound to active human neurokinin 2 receptor in complex with G324 | | Descriptor: | G324, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W.J, Yuan, Q.N, Zhang, H.H, Yang, F, Ling, S.L, Lv, P, Eric, X, Tian, C.L, Yin, W.C, Shi, P. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the activation of neurokinin 2 receptor by neurokinin A.
Cell Discov, 8, 2022
|
|
7S83
 
 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | Authors: | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
