7JVC
 
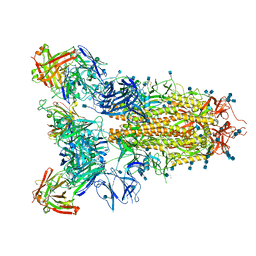 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV4
 
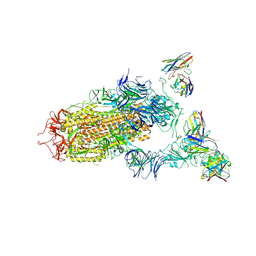 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | Authors: | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-08-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7K3Q
 
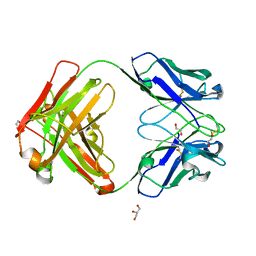 | | An ultra-potent human neutralizing antibody locks the SARS-CoV-2 spike in the closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Fab fragment of S2E12 monoclonal antibody, heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Ng, C. | | Deposit date: | 2020-09-12 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms.
Science, 370, 2020
|
|
6UMM
 
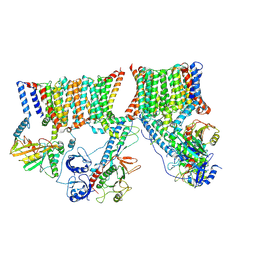 | | A complete structure of the ESX-3 translocon complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Poweleit, N, Rosenberg, O.S. | | Deposit date: | 2019-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A large inner membrane pore defines the ESX translocon
To Be Published
|
|
8S9G
 
 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7K45
 
 | |
7K4N
 
 | |
7R7N
 
 | |
7RA8
 
 | |
7RAL
 
 | |
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7TLY
 
 | |
7TM0
 
 | |
7TLZ
 
 | |
9ASD
 
 | | VIR-7229 Fab fragment bound the SARS-CoV-2 BA.2.86 spike trimer (local refinement of the BA 2.86 RBD/VIR-7229 VHVL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VIR-7229 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9AU2
 
 | | VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Park, Y.J, Veelser, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
8G30
 
 | |
8G3Q
 
 | |
8G3R
 
 | |
8G3O
 
 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G40
 
 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI19 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G3M
 
 | |
8G3N
 
 | | N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
