4AD3
 
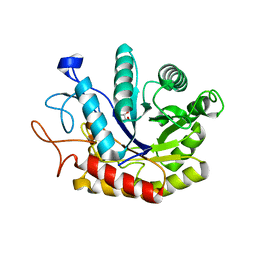 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ACZ
 
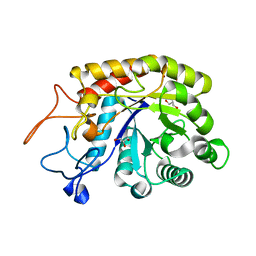 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD5
 
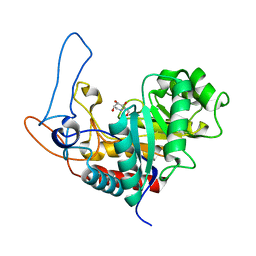 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-deoxymannojirimycin and alpha-1,2-mannobiose | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD2
 
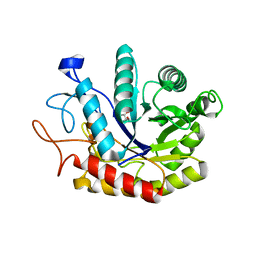 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD0
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BM7
 
 | | Crystal Structure of IgG Fc F241A mutant with native glycosylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, IG GAMMA-1 CHAIN C REGION | | Authors: | Yu, X, Baruah, K, Harvey, D.J, Vasiljevic, S, Alonzi, D.S, Song, B, Higgins, M.K, Bowden, T.A, Crispin, M, Scanlan, C.N. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Engineering Hydrophobic Protein-Carbohydrate Interactions to Fine-Tune Monoclonal Antibodies.
J.Am.Chem.Soc., 135, 2013
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.106 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
7MFK
 
 | |
7MFL
 
 | |
6FSN
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
5MHF
 
 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-916a2c5a-4 (Mpro-x3303) | | Descriptor: | 3C-like proteinase, 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | Descriptor: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-4 (Mpro-x10566) | | Descriptor: | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCT
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-LEF-c49414a7-1 (Mpro-x10756) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with SIM-SYN-f15aaa3a-1 (Mpro-x3348) | | Descriptor: | 1-[4-(diphenylmethyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAR-DIA-eace69ff-36 (Mpro-x3351) | | Descriptor: | 1-(5-fluoro-1H-indol-3-yl)-N-methylmethanamine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD1
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-58 (Mpro-x10856) | | Descriptor: | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UNI-f8e79267-2 (Mpro-x10889) | | Descriptor: | (2R)-4-[(methanesulfonyl)amino]-2-phenyl-N-(pyridin-3-yl)butanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with GAB-REV-70cc3ca5-18 (Mpro-x3366) | | Descriptor: | 2-(1H-benzimidazol-6-yl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
