4SKN
 
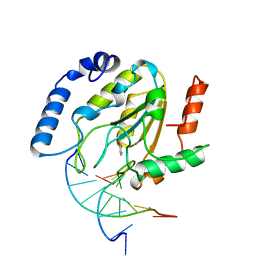 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | Authors: | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-02-20 | | Release date: | 1999-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
4TWS
 
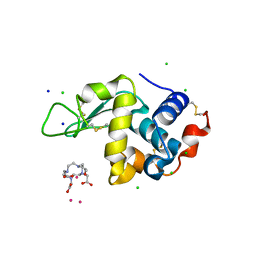 | | Gadolinium Derivative of Tetragonal Hen Egg-White Lysozyme at 1.45 A Resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Holton, J.M, Classen, S, Frankel, K.A, Tainer, J.A. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures.
Febs J., 281, 2014
|
|
5UM9
 
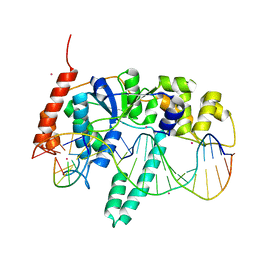 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
1T38
 
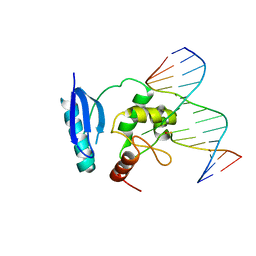 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE BOUND TO DNA CONTAINING O6-METHYLGUANINE | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(6OG)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T39
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE COVALENTLY CROSSLINKED TO DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(E1X)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3L41
 
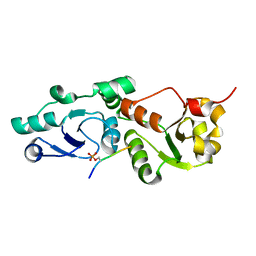 | | Crystal Structure of S. pombe Brc1 BRCT5-BRCT6 domains in complex with phosphorylated H2A | | Descriptor: | BRCT-containing protein 1, phosphorylated H2A tail | | Authors: | Williams, R.S, Williams, J.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | gammaH2A binds Brc1 to maintain genome integrity during S-phase.
Embo J., 29, 2010
|
|
6X1Y
 
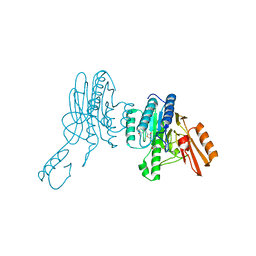 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
3L2P
 
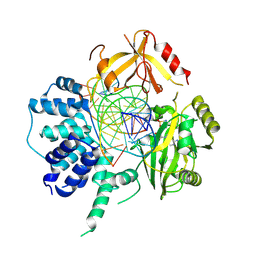 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
6WFV
 
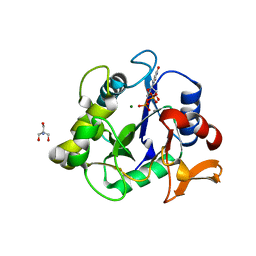 | | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Guo, H.-F, Tsai, C.-L, Miller, M.D, Phillips Jr, G.N, Tainer, J.A, Kurie, J.M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human
To Be Published
|
|
3L40
 
 | |
6WH2
 
 | | Structure of the C-terminal BRCT domain of human XRCC1 | | Descriptor: | X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
6X1Z
 
 | | Mre11 dimer in complex with small molecule modulator PFMJ | | Descriptor: | (5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
2NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND WATER BOUND IN ACTIVE CENTER | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
5K97
 
 | | Flap endonuclease 1 (FEN1) D233N with cleaved product fragment and Sm3+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
5KSE
 
 | | Flap endonuclease 1 (FEN1) R100A with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2016-07-08 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
6VBH
 
 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VBA
 
 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6WH1
 
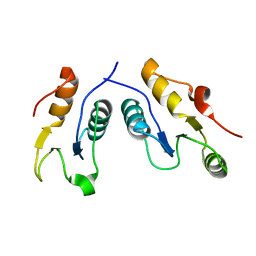 | | Structure of the complex of human DNA ligase III-alpha and XRCC1 BRCT domains | | Descriptor: | DNA ligase 3 alpha, X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
8D3O
 
 | |
8D3G
 
 | |
8D3H
 
 | |
8D3J
 
 | |
8D3E
 
 | |
8D3N
 
 | |
8D3I
 
 | |
