4DDR
 
 | | Human dihydrofolate reductase complexed with NADPH and P218 | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yuthavong, Y, Tarnchompoo, B, Vilaivan, T, Chitnumsub, P, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Thongphanchang, C, Taweechai, S, Vanichtanankul, J, Rattanajak, R, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DPH
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P65 and NADPH | | Descriptor: | 2,4-diamino-6-methyl-5-[3-(2,4,5-trichlorophenoxy)propyloxy]pyrimidine, BETA-MERCAPTOETHANOL, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5K9A
 
 | | Sortase A from Corynebacterium diphtheriae | | Descriptor: | Putative fimbrial associated sortase-like protein, SULFATE ION | | Authors: | Osipiuk, J, Huang, I.-H, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
3IRU
 
 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | Descriptor: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6NZH
 
 | |
6NZF
 
 | |
6NZE
 
 | |
4WOV
 
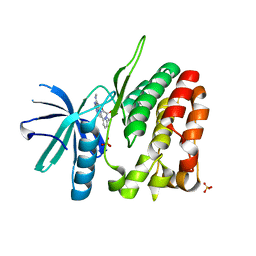 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH BMS-066 AKA 2-METHOXY-N-({6-[3-METHYL-7-(METHYLAMINO)-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0, 6]DODECA-1(9),2(6),4,7,11-PENTAEN-11-YL]PYRIDIN-2-YL}METHY L)ACETAMIDE | | Descriptor: | 2-methoxy-N-({6-[1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl]pyridin-2-yl}methyl)acetamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2014-10-16 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine Kinase 2-mediated Signal Transduction in T Lymphocytes Is Blocked by Pharmacological Stabilization of Its Pseudokinase Domain.
J.Biol.Chem., 290, 2015
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4GFM
 
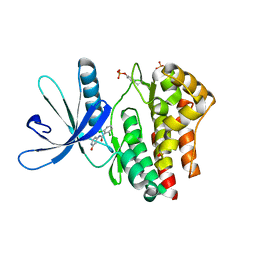 | | JAK2 kinase (JH1 domain) with 2,6-DICHLORO-N-(2-OXO-2,5-DIHYDROPYRIDIN-4-YL)BENZAMIDE | | Descriptor: | 2,6-dichloro-N-(2-oxo-2,5-dihydropyridin-4-yl)benzamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4GIH
 
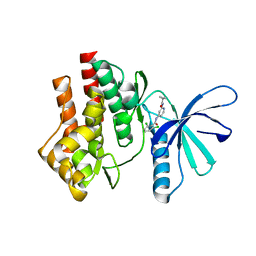 | |
5WBW
 
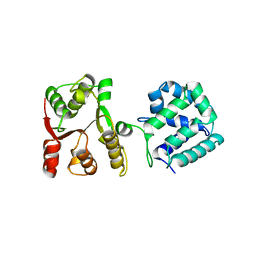 | | Yeast Hsp104 fragment 1-360 | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.
Biosci. Rep., 37, 2017
|
|
4GMY
 
 | |
4GDY
 
 | | Kynurenine Aminotransferase II inhibitors | | Descriptor: | (5-hydroxy-6-methyl-4-{[(1-oxo-7-phenoxy-1,2-dihydro[1,2,4]triazolo[4,3-a]quinolin-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of Hydroxamate Bioisosteres as KATII Inhibitors with Improved Oral Bioavailability and Pharmacokinetics
MEDCHEMCOMM, 2012
|
|
4GE9
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (4-{[(6-benzyl-1-hydroxy-7-methoxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Design of Irreversible Human KAT II Inhibitors: Discovery of New Potency-Enhancing Interactions.
ACS Med Chem Lett, 4, 2013
|
|
6NZ4
 
 | | YcjX-GDP (type I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
7MU2
 
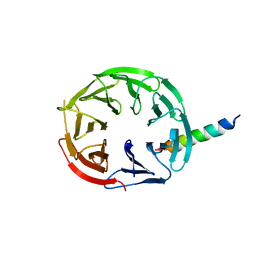 | | Crystal Structure of WIPI2 in complex with W2IR of ATG16L1 | | Descriptor: | Autophagy-related protein 16-1, WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Strong, L.M, Flower, T.G, Buffalo, C.Z, Hurley, J.H. | | Deposit date: | 2021-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy.
Elife, 10, 2021
|
|
4W9T
 
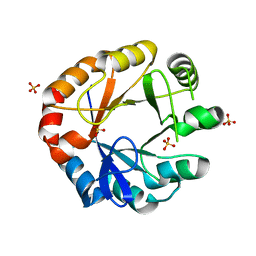 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4WUI
 
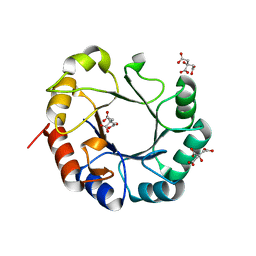 | | Crystal structure of TrpF from Jonesia denitrificans | | Descriptor: | CITRIC ACID, N-(5'-phosphoribosyl)anthranilate isomerase | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
6RJK
 
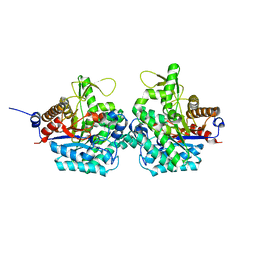 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | Descriptor: | Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJO
 
 | | Complex structure of virulence factor SghA with its substrate analog salicin | | Descriptor: | 2-(hydroxymethyl)phenyl beta-D-glucopyranoside, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-28 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RK2
 
 | | Complex structure of virulence factor SghA mutant with its substrate SAG | | Descriptor: | 2-(alpha-L-altropyranosyloxy)benzoic acid, Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJM
 
 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | Descriptor: | Beta-glucosidase, alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
