7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
6LJB
 
 | |
1L2J
 
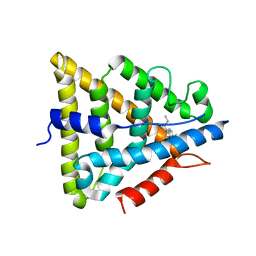 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, ESTROGEN RECEPTOR BETA | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
6LJ9
 
 | |
6LNQ
 
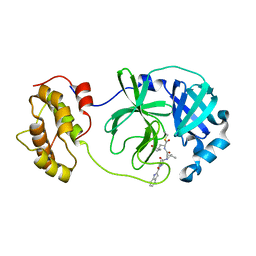 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | Descriptor: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | Authors: | Wang, H, Shang, L.Q. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
6LO0
 
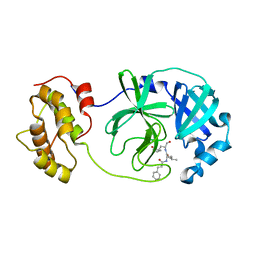 | |
8XK5
 
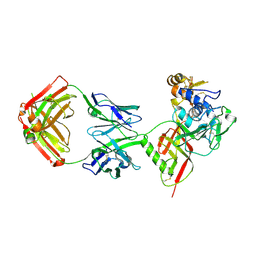 | | SNB1G11 Fab bound to SFTSV glycoprotein Gn | | Descriptor: | Envelopment polyprotein, mAb SNB1G11 Fab heavy chain, mAb SNB1G11 Fab light chain | | Authors: | Deng, Z. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A broadly protective antibody targeting glycoprotein Gn inhibits severe fever with thrombocytopenia syndrome virus infection.
Nat Commun, 15, 2024
|
|
8XK8
 
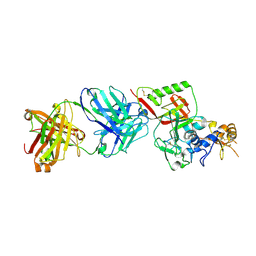 | | N1D10 Fab bound to SFTSV glycoprotein-Gn | | Descriptor: | Envelopment polyprotein, mAb N1D10 Fab heavy chain, mAb N1D10 Fab light chain | | Authors: | Zhao, H, Deng, Z. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | A broadly protective antibody targeting glycoprotein Gn inhibits severe fever with thrombocytopenia syndrome virus infection.
Nat Commun, 15, 2024
|
|
8XK6
 
 | | S2A5 Fab bound to SFTSV glycoprotein Gn | | Descriptor: | Envelopment polyprotein, mAb S2A5 Fab heavy chain, mAb S2A5 Fab light chain | | Authors: | Deng, Z. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A broadly protective antibody targeting glycoprotein Gn inhibits severe fever with thrombocytopenia syndrome virus infection.
Nat Commun, 15, 2024
|
|
6LNY
 
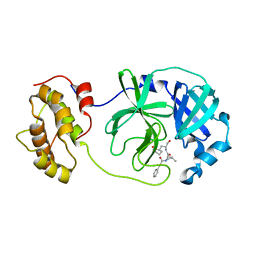 | |
2FV5
 
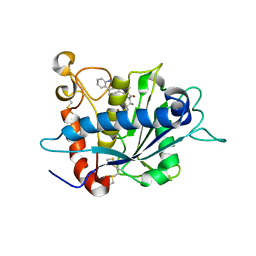 | | Crystal structure of TACE in complex with IK682 | | Descriptor: | (2R)-N-HYDROXY-2-[(3S)-3-METHYL-3-{4-[(2-METHYLQUINOLIN-4-YL)METHOXY]PHENYL}-2-OXOPYRROLIDIN-1-YL]PROPANAMIDE, ADAM 17, ZINC ION | | Authors: | Orth, P, Niu, X. | | Deposit date: | 2006-01-30 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IK682, a tight binding inhibitor of TACE.
Arch.Biochem.Biophys., 451, 2006
|
|
7CMZ
 
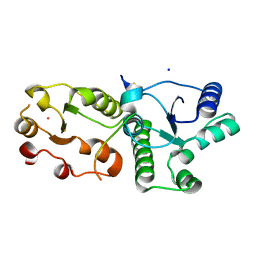 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | Authors: | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
8JF4
 
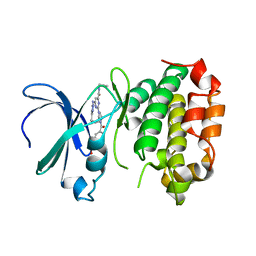 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | Authors: | Zhu, C.J. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89288354 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JF3
 
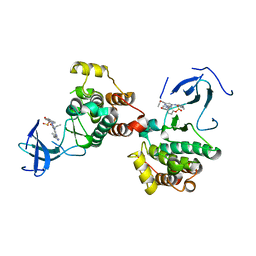 | | C-Src in complex with compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhang, Z.M, Huang, H.S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.84647632 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8INB
 
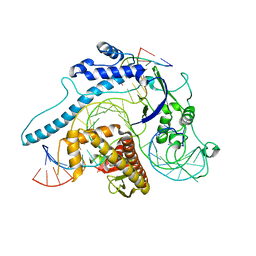 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | Descriptor: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
8JG8
 
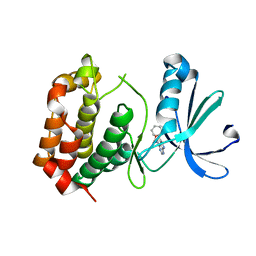 | |
2JR7
 
 | | Solution structure of human DESR1 | | Descriptor: | DPH3 homolog, ZINC ION | | Authors: | Wu, F, Wu, J, Shi, Y. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human DESR1, a CSL zinc-binding protein
Proteins, 71, 2008
|
|
7X98
 
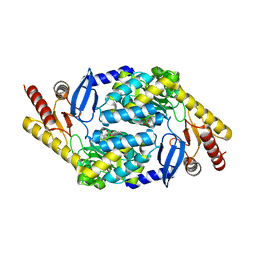 | |
7VV9
 
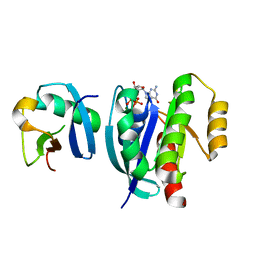 | |
7VVG
 
 | |
7VVB
 
 | |
7VV8
 
 | |
2CAY
 
 | | Vps36 N-terminal PH domain | | Descriptor: | SULFATE ION, VACUOLAR PROTEIN SORTING PROTEIN 36 | | Authors: | Teo, H, Williams, R.L, Perisic, O, Gill, D.J. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
7YSU
 
 | | Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
