4CIC
 
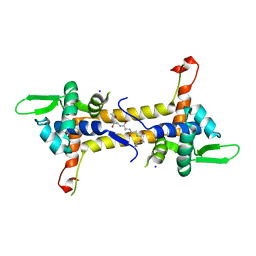 | | T. potens IscR | | Descriptor: | HEXA-ALANINE PEPTIDE, SODIUM ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Santos, J.A, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Unique Regulation of Iron-Sulfur Cluster Biogenesis in a Gram-Positive Bacterium.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4EZF
 
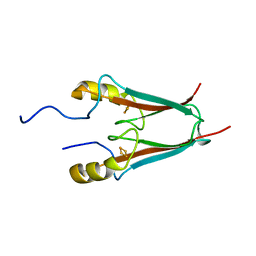 | | The Crystal Structure of a Human MitoNEET mutant with an Ala inserted between Asp 67 and Lys 68 | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Baxter, E.L, Zuris, J.A, Wang, C, Axelrod, H.L, Cohen, A.E, Paddock, M.L, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2012-05-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Allosteric control in a metalloprotein dramatically alters function.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DE8
 
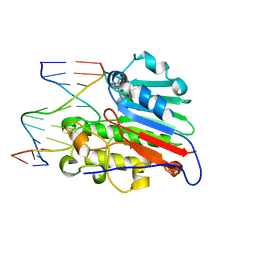 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1NAT
 
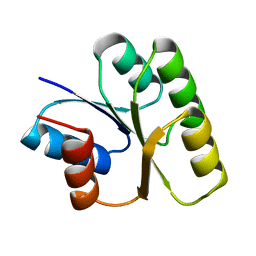 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
1ZWX
 
 | | Crystal Structure of SmcL | | Descriptor: | GLYCEROL, PHOSPHATE ION, sphingomyelinase-c | | Authors: | Openshaw, A.E.A, Race, P.R, Monzo, H.J, Vasquez-Boland, J.A, Banfield, M.J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria.
J.Biol.Chem., 280, 2005
|
|
2Y1W
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | 2-{4-[3-FLUORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL] PIPERIDIN-1-YL}-N-METHYLETHANAMINE, HISTONE-ARGININE METHYLTRANSFERASE CARM1, SINEFUNGIN | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
151D
 
 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
4F71
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, wild-type protein, complex with magnesium and inorganic phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
2XYZ
 
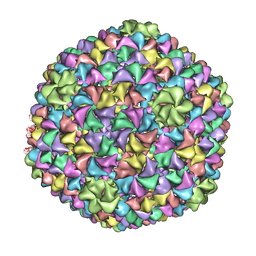 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2AA5
 
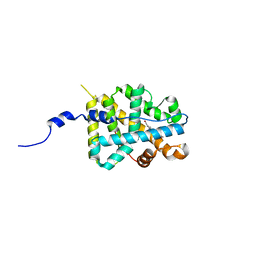 | | Mineralocorticoid Receptor with Bound Progesterone | | Descriptor: | Mineralocorticoid receptor, PROGESTERONE | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
2XD4
 
 | |
4F8M
 
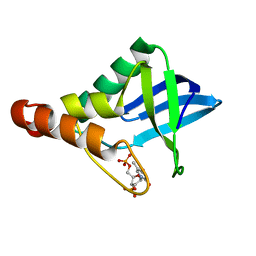 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23I/I92V at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-05-17 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
1AKZ
 
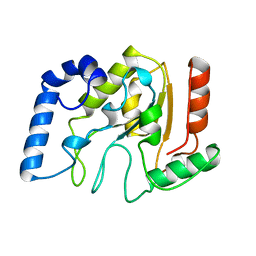 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
2A8I
 
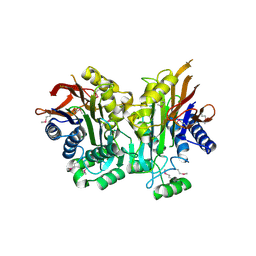 | | Crystal Structure of human Taspase1 | | Descriptor: | Threonine aspartase 1 | | Authors: | Khan, J.A, Dunn, B.M, Tong, L. | | Deposit date: | 2005-07-08 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Taspase1, a Crucial Protease Regulating the Function of MLL.
Structure, 13, 2005
|
|
4F1E
 
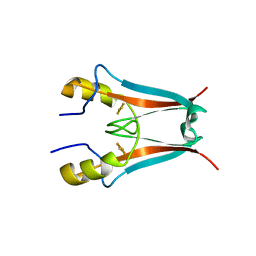 | | The Crystal Structure of a Human MitoNEET mutant with Asp 67 replaced by a Gly | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Baxter, E.I, Zuris, J.A, Wang, C, Axelrod, H.L, Cohen, A.E, Paddock, M.L, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2012-05-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric control in a metalloprotein dramatically alters function.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2AAX
 
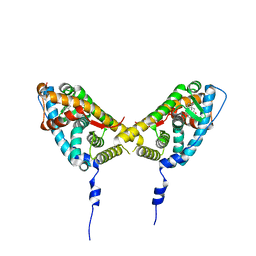 | | Mineralocorticoid Receptor Double Mutant with Bound Cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, Mineralocorticoid receptor, SULFATE ION | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
1A1Q
 
 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
2XRU
 
 | | AURORA-A T288E COMPLEXED WITH PHA-828300 | | Descriptor: | 3-({[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]CARBONYL}AMINO)-N-[(1R)-1-PHENYLPROPYL]-1H-THIENO[3,2-C]PYRAZOLE-5-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Bindi, S, Fancelli, D, Alli, C, Berta, D, Bertrand, J.A, Cameron, A.D, Cappella, P, Carpinelli, P, Cervi, G, Croci, W, D'Anello, M, Forte, B, LauraGiorgini, M, Marsiglio, A, Moll, J, Pesenti, E, Pittala, V, Pulici, M, Riccardi-Sirtori, F, Roletto, F, Soncini, C, Storici, P, Varasi, M, Volpi, D, Zugnoni, P, Vianello, P. | | Deposit date: | 2010-09-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bioorg.Med.Chem., 18, 2010
|
|
1Q97
 
 | | The structure of the Saccharomyces cerevisiae SR protein kinase, Sky1p, with bound ATP | | Descriptor: | ADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nolen, B, Ngo, J, Chakrabarti, S, Vu, D, Adams, J.A, Ghosh, G. | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide-Induced Conformational Changes in the Saccharomyces cerevisiae SR Protein Kinase, Sky1p, Revealed by X-ray Crystallography
Biochemistry, 42, 2003
|
|
4F7X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
2Y35
 
 | | Crystal structure of Xrn1-substrate complex | | Descriptor: | DT11 (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', LD22664P, MAGNESIUM ION | | Authors: | Jinek, M, Coyle, S.M, Doudna, J.A. | | Deposit date: | 2010-12-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coupled 5' Nucleotide Recognition and Processivity in Xrn1-Mediated Mrna Decay.
Mol.Cell, 41, 2011
|
|
2A3A
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline | | Descriptor: | SULFATE ION, THEOPHYLLINE, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
