5MLA
 
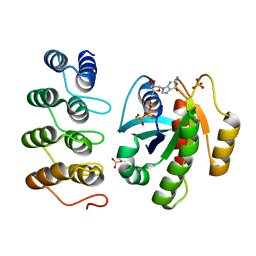 | | Crystal structure of human RAS in complex with darpin K55 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Inhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
5MLB
 
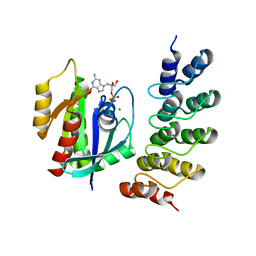 | | Crystal structure of human RAS in complex with darpin K27 | | Descriptor: | DARPin K27, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | INhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
7EXT
 
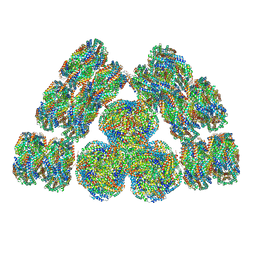 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7EYD
 
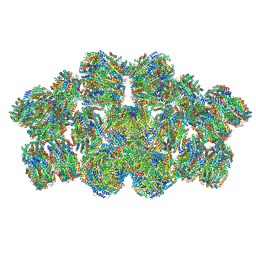 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | Descriptor: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-30 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
6PMO
 
 | |
6POM
 
 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6QRZ
 
 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
6WXK
 
 | | PHF23 PHD Domain Apo | | Descriptor: | PHD finger protein 23, ZINC ION | | Authors: | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
6JZO
 
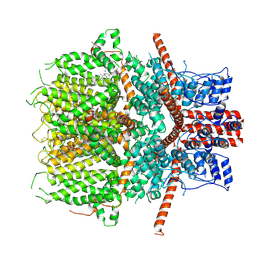 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-10-21 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel
To Be Published
|
|
6P05
 
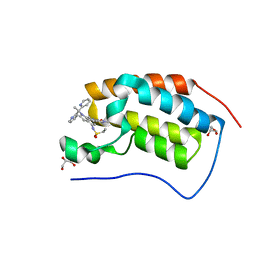 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with compound 27 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Zhao, J, Gutgesell, L.M, Shen, Z, Dye, K, Dubrovyskyii, O, Zhao, H, Huang, F, Tonetti, D.A, Thatcher, G.R. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7BER
 
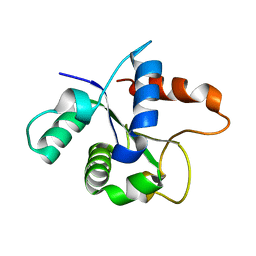 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7BEQ
 
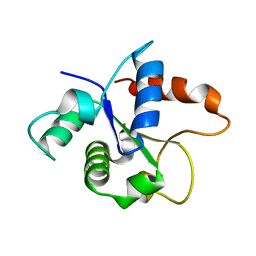 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
4APP
 
 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
3IZ3
 
 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5O2S
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
5O2T
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
6JJP
 
 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
2DK9
 
 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
6XBU
 
 | | polymerase domain of polymerase-theta | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*TP*CP*AP*TP*TP*G)-3'), DNA polymerase theta, ... | | Authors: | Chen, X, Pomerantz, R, Zhao, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pol theta reverse transcribes RNA and promotes RNA-templated DNA repair.
Sci Adv, 7, 2021
|
|
1TOV
 
 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | Descriptor: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | Authors: | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
6ZRK
 
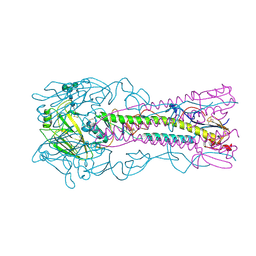 | | Crystal structure of H8 haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Xiong, X, Walker, P, Zhang, J, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-07-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hemagglutinin Structure and Activities.
Cold Spring Harb Perspect Med, 11, 2021
|
|
7A9D
 
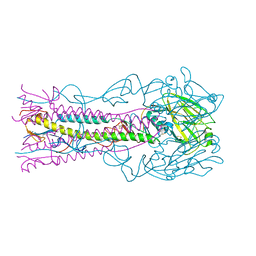 | | Crystal structure of H12 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Xiong, X, Walker, P, Zhang, J, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hemagglutinin Structure and Activities.
Cold Spring Harb Perspect Med, 11, 2021
|
|
2GIB
 
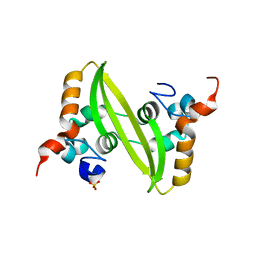 | | Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain | | Descriptor: | Nucleocapsid protein, SULFATE ION | | Authors: | Yu, I.M, Oldham, M.L, Zhang, J, Chen, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae.
J.Biol.Chem., 281, 2006
|
|
