5TTO
 
 | | X-ray crystal structure of PPARgamma in complex with SR1643 | | Descriptor: | 4-bromo-N-{3,5-dichloro-4-[(quinolin-3-yl)oxy]phenyl}-2,5-difluorobenzene-1-sulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P, Kamenecka, T, Abell, A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPAR gamma-Targeted Antidiabetics.
J. Med. Chem., 60, 2017
|
|
4MQ2
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MQ1
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-(5-{[(1R)-3-amino-1-(3-chlorophenyl)propyl]carbamoyl}-2-chlorophenyl)-2-methoxy-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, PENTAETHYLENE GLYCOL, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4OEW
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
5JA5
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JHP
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
7LKI
 
 | |
5J9K
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, ZINC ION, rice D53 peptide 794-808 | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JGC
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A, L130A, L179A and I195A mutant | | Descriptor: | Protein TPR1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-20 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
7CSW
 
 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
5H4B
 
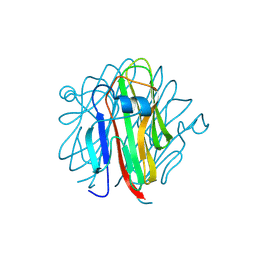 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
8SZX
 
 | |
8Y0Q
 
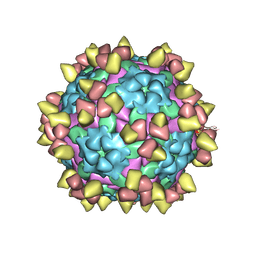 | | Complex of FMDV O/18074 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Discovery, recognized antigenic structures, and evolution of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus.
Plos Pathog., 20, 2024
|
|
8Y0R
 
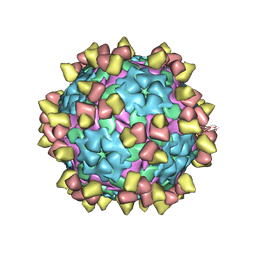 | | Complex of FMDV A/WH/CHA/09 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Discovery, recognized antigenic structures, and evolution of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus.
Plos Pathog., 20, 2024
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
5U2N
 
 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
5U6D
 
 | |
6L8H
 
 | | Crystal structure of CYP97C1 | | Descriptor: | Carotene epsilon-monooxygenase, chloroplastic, GLYCEROL, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5GHZ
 
 | |
4HVI
 
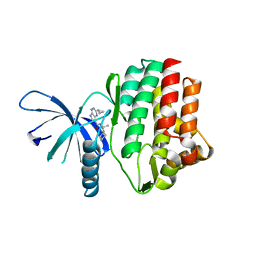 | |
5GHX
 
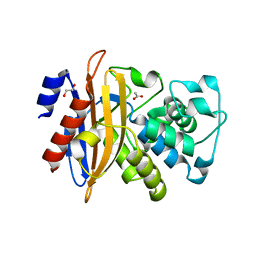 | | Crystal structure of beta-lactamase PenP mutant-E166H | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2016-06-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Snapshots of Class A beta-Lactamase Catalysis Reveal Structural Changes That Facilitate beta-Lactam Hydrolysis
J. Biol. Chem., 292, 2017
|
|
6L8J
 
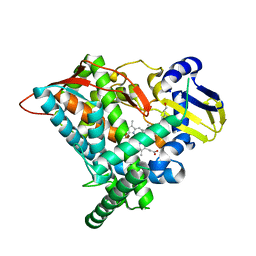 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | Authors: | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | Deposit date: | 2017-10-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
