7OEH
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ059b | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3S)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OEG
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ040b | | Descriptor: | (R)-4-((2-azaspiro[3.3]heptan-2-yl)methyl)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OEM
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ120 | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-methylsulfanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
6RT5
 
 | |
6RT7
 
 | |
6RT6
 
 | |
6SQM
 
 | | Crystal structure of CREBBP bromodomain complexed with LA36 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-acetamido-5-[(3-methylcinnolin-5-yl)carbamoyl]phenyl]furan-2-carboxamide | | Authors: | Bedi, R.K, Laul, E, Nevado, C, Caflisch, A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexed with LA36
To Be Published
|
|
6RT4
 
 | |
6SXX
 
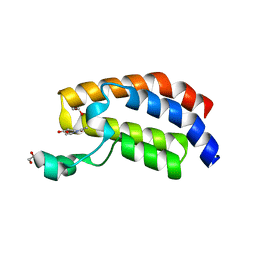 | | Crystal structure of the bromodomain of human CREBBP in complex with ACA007 | | Descriptor: | (3~{R})-1-ethanoylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREBBP | | Authors: | Xiang, W, Bedi, R.K, Sledz, P, Caflisch, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with ACA007
To Be Published
|
|
6SQE
 
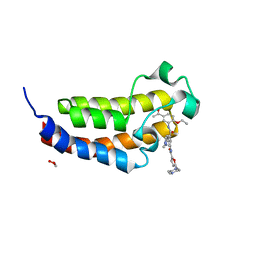 | | Crystal structure of CREBBP bromodomain complexed with KD341 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]-5-(1-methylpyrazol-3-yl)phenyl]-5-[(4-methylpiperazin-1-yl)methyl]furan-2-carboxamide | | Authors: | Bedi, R.K, Kirillova, M, Nevado, C, Caflisch, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexed with KD341
To Be Published
|
|
6SQF
 
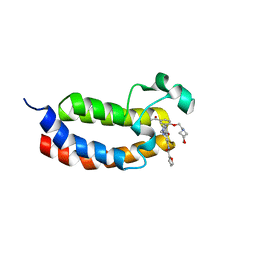 | |
6SW3
 
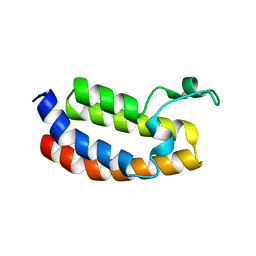 | |
2RU4
 
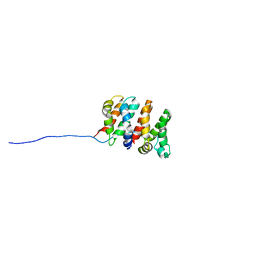 | | Designed Armadillo Repeat Protein Self-ASsembled Complex (YIIM2-MAII) | | Descriptor: | Armadillo Repeat Protein, C-terminal fragment, MAII, ... | | Authors: | Zerbe, O, Christen, M.T, Plueckthun, A, Watson, R.P. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure
Structure, 22, 2014
|
|
2RU5
 
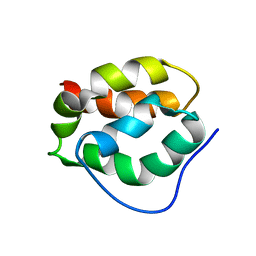 | |
5J94
 
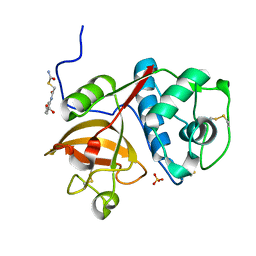 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC13345 | | Descriptor: | 2-{[(carbamoylsulfanyl)acetyl]amino}benzoic acid, Cathepsin K, SULFATE ION | | Authors: | Novinec, M, Korenc, M, Lenarcic, B, Baici, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22002459 Å) | | Cite: | A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods.
Nat Commun, 5, 2014
|
|
6Y86
 
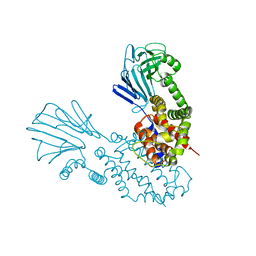 | |
2XYH
 
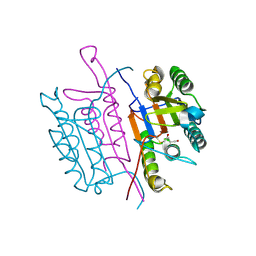 | | Caspase-3:CAS60254719 | | Descriptor: | 5-CHLORO-4-OXOPENTANOIC ACID, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XYG
 
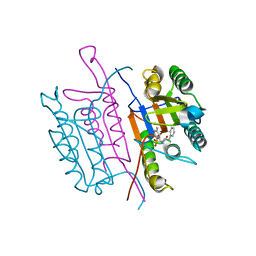 | | Caspase-3:CAS329306 | | Descriptor: | CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XYP
 
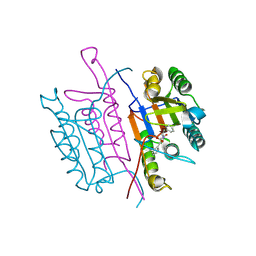 | | Caspase-3:CAS26049945 | | Descriptor: | CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17, PHENYLMETHYL N-[(2S)-4-CHLORO-3-OXO-1-PHENYL-BUTAN-2-YL]CARBAMATE | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-18 | | Release date: | 2011-08-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2M10
 
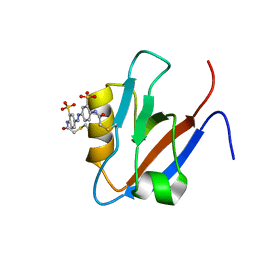 | |
2M0Z
 
 | | cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Walser, R, Zerbe, O, Hamm, P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-03 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Kinetic response of a photoperturbed allosteric protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CPS
 
 | |
6EPS
 
 | | The ATAD2 bromodomain in complex with compound UZH-DQ41 | | Descriptor: | (2~{R})-~{N}-[5-[3,5-bis(oxidanyl)phenyl]-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Hitting a moving target: simulation and crystallography study of ATAD2 bromodomain blockers
To Be Published
|
|
8QAN
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with acetyl-pyrrole derivative compound 79 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-methyl-4-(3-methylbutyl)-5-(2-piperazin-1-yl-1,3-thiazol-4-yl)-1~{H}-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Enhanced cellular death in liver and breast cancer cells by dual BET/BRPF1 inhibitors.
Protein Sci., 33, 2024
|
|
8QAP
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with acetyl-pyrrole derivative compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(3,6,6-trimethyl-4-oxidanylidene-5,7-dihydro-2~{H}-isoindol-1-yl)-1,3-thiazol-2-yl]guanidine, Bromodomain-containing protein 4 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced cellular death in liver and breast cancer cells by dual BET/BRPF1 inhibitors.
Protein Sci., 33, 2024
|
|
