6V5D
 
 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
4V3R
 
 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | MAGNESIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1W5D
 
 | | Crystal structure of PBP4a from Bacillus subtilis | | Descriptor: | CALCIUM ION, PENICILLIN-BINDING PROTEIN | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-06 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Penicillin-Binding Protein 4A, and its Complex with a Peptidoglycan Mimetic Peptide.
J.Mol.Biol., 371, 2007
|
|
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
9F5H
 
 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
4V8N
 
 | | The crystal structure of agmatidine tRNA-Ile2 bound to the 70S ribosome in the A and P site. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Mandal, D, Neubauer, C, Koehrer, C, RajBhandary, U.L, Ramakrishnan, V. | | Deposit date: | 2013-02-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structural Basis for Specific Decoding of Aua by Isoleucine tRNA on the Ribosome
Nat.Struct.Mol.Biol., 20, 2013
|
|
1WHO
 
 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
4WBC
 
 | | 2.13 A STRUCTURE OF A KUNITZ-TYPE WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN | | Descriptor: | PROTEIN (CHYMOTRYPSIN INHIBITOR), SULFATE ION | | Authors: | Ravichandran, S, Sen, U, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Cryocrystallography of a Kunitz-type serine protease inhibitor: the 90 K structure of winged bean chymotrypsin inhibitor (WCI) at 2.13 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2QLQ
 
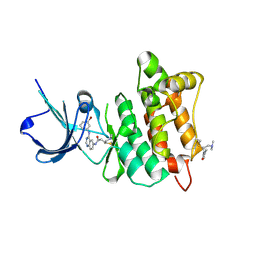 | | Crystal structure of SRC kinase domain with covalent inhibitor RL3 | | Descriptor: | (2E)-N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
4UYB
 
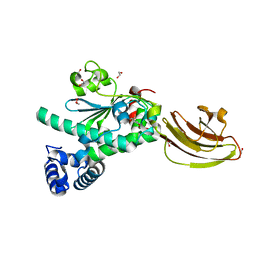 | | Crystal structure of SEC14-like protein 3 | | Descriptor: | 1,2-ETHANEDIOL, SEC14-LIKE PROTEIN 3, UNKNOWN LIGAND | | Authors: | Kopec, J, Goubin, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Sec14-Like Protein 3
To be Published
|
|
2GA2
 
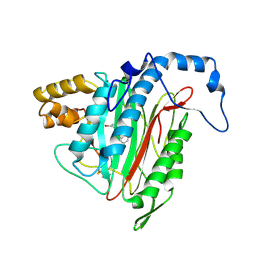 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2QI8
 
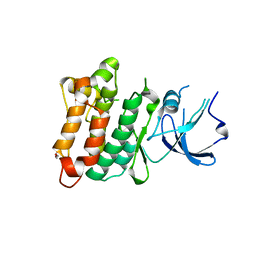 | | Crystal structure of drug resistant SRC kinase domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
2G9Z
 
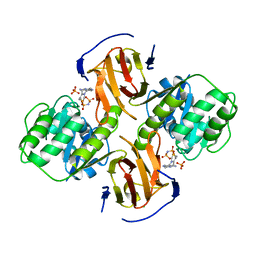 | | Thiamin pyrophosphokinase from Candida albicans | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-(2-{[HYDROXY(PHOSPHONOAMINO)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-I UM, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Abergel, C, Santini, S, Monchois, V, Rousselle, T, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2006-03-07 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Bmc Struct.Biol., 8, 2008
|
|
8PJ0
 
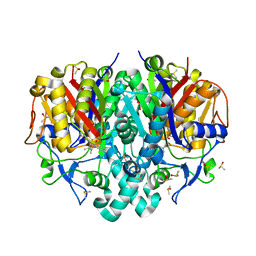 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-3-methylbutanamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8P5Q
 
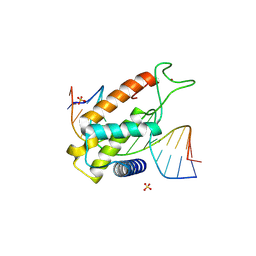 | | Structure of an ALOG domain from Arabidopsis thaliana in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*CP*AP*AP*CP*AP*GP*TP*AP*AP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*TP*TP*AP*CP*TP*GP*TP*TP*GP*AP*CP*GP*T)-3'), Protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 3, ... | | Authors: | Zubieta, C, Nanao, M.H, Rieu, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The ALOG domain defines a family of plant-specific transcription factors acting during Arabidopsis flower development.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4URU
 
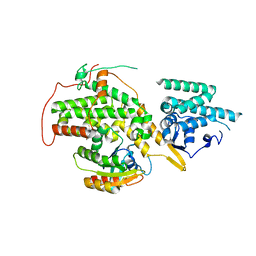 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 4-METHOXY-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
2QQ7
 
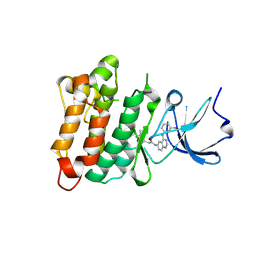 | | Crystal structure of drug resistant SRC kinase domain with irreversible inhibitor | | Descriptor: | (2E)-N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-26 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
4V0G
 
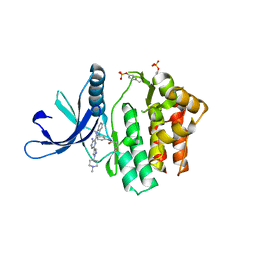 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
9F8H
 
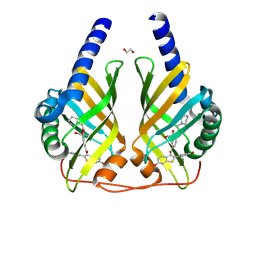 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with Raloxifene | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phenazine biosynthesis protein A/B, ... | | Authors: | Diederich, C, Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
4WIW
 
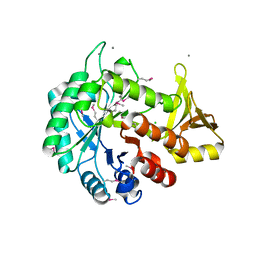 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | Authors: | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
4WHI
 
 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | Descriptor: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
4KUM
 
 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
