8Y8D
 
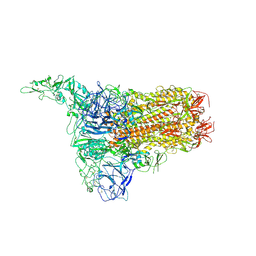 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8F
 
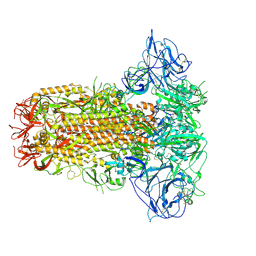 | | Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8I
 
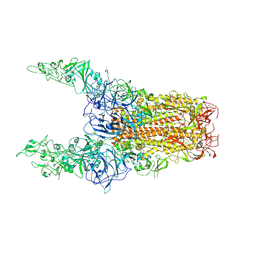 | | Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8J
 
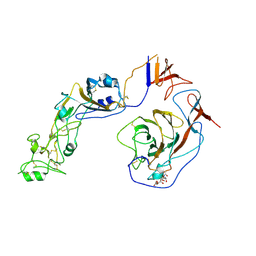 | | Local structure of HCoV-HKU1C spike in complex with glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8C
 
 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8G
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8H
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
3H2L
 
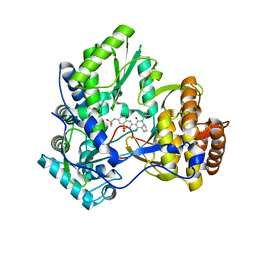 | | Crystal structure of HCV NS5B polymerase in complex with a novel bicyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(4aR,7aS)-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,4a,5,6,7,7a-hexahydro-1H-cyclopenta[b]pyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, NS5B polymerase | | Authors: | Han, Q, Showalter, R.E, Zhou, Q, Kissinger, C.R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of tricyclic 5,6-dihydro-1H-pyridin-2-ones as novel, potent, and orally bioavailable inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1BJJ
 
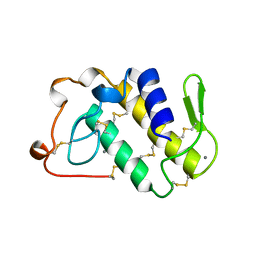 | | AGKISTRODOTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | Descriptor: | AGKISTRODOTOXIN, CALCIUM ION | | Authors: | Tang, L, Zhou, Y, Lin, Z. | | Deposit date: | 1998-06-25 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of agkistrodotoxin in an orthorhombic crystal form with six molecules per asymmetric unit.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3EWH
 
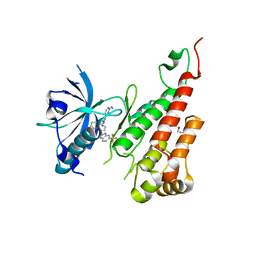 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridyl-pyrimidine benzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-6-(trifluoromethyl)-1H-benzimidazol-2-amine, vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPY
 
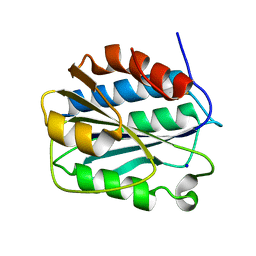 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
5KZI
 
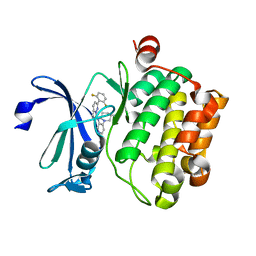 | | Crystal structure of human Pim-1 kinase in complex with an imidazopyridazine inhibitor. | | Descriptor: | Serine/threonine-protein kinase pim-1, ~{N}-[4-[(3~{S})-3-azanylpiperidin-1-yl]pyridin-3-yl]-2-[2,6-bis(fluoranyl)phenyl]imidazo[1,5-b]pyridazin-7-amine | | Authors: | Mohr, C. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of imidazopyridazines as potent Pim-1/2 kinase inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
3PPW
 
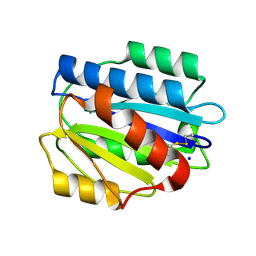 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
5JIE
 
 | |
3LQQ
 
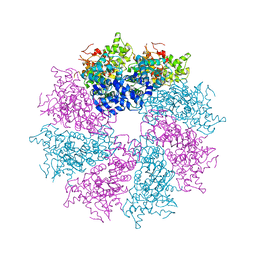 | | Structure of the CED-4 Apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.534 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LQR
 
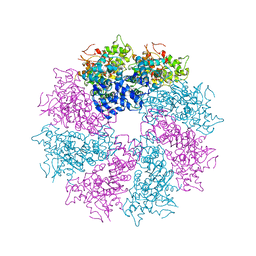 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
1GH2
 
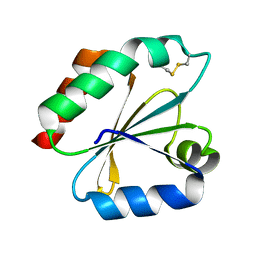 | | Crystal structure of the catalytic domain of a new human thioredoxin-like protein | | Descriptor: | THIOREDOXIN-LIKE PROTEIN | | Authors: | Jin, J, Chen, X, Guo, Q, Yuan, J, Qiang, B, Rao, Z. | | Deposit date: | 2000-11-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the catalytic domain of a human thioredoxin-like protein.
Eur.J.Biochem., 269, 2002
|
|
7KLZ
 
 | |
5ZKP
 
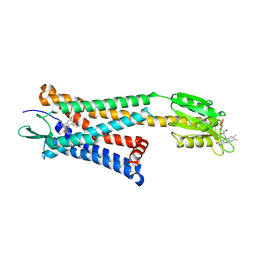 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8Y22
 
 | | FGFR1 kinase domain with a covalent inhibitor 9g | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}-[4-[[4-azanyl-3-(7-methoxy-5-methyl-1-benzothiophen-2-yl)pyrazolo[3,4-d]pyrimidin-1-yl]methyl]phenyl]propanamide | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
8XZ7
 
 | | FGFR1 kinase domain with a covalent inhibitor 10h | | Descriptor: | 5-azanyl-3-[2-[4,6-bis(fluoranyl)-2-methyl-3~{H}-benzimidazol-5-yl]ethynyl]-1-[[3-(prop-2-enoylamino)phenyl]methyl]pyrazole-4-carboxamide, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
