7VUJ
 
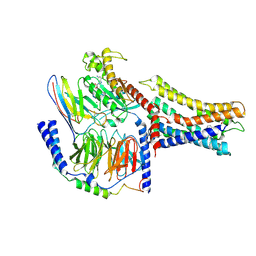 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUG
 
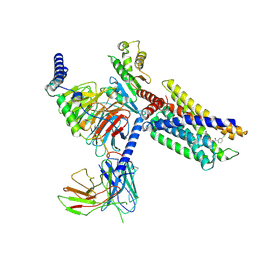 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
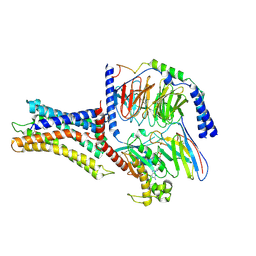 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7D9Q
 
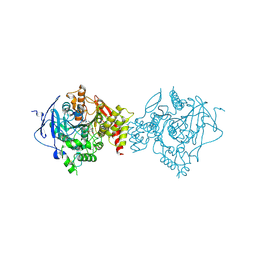 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 7 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(3-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9P
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 12 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(2-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9O
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | Descriptor: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM5
 
 | |
7VM6
 
 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM7
 
 | | Crystal structure of inactive uPA in complex with nafamostat | | Descriptor: | (6-carbamimidoylnaphthalen-2-yl) 4-carbamimidamidobenzoate, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
5T2P
 
 | | Hepatitis B virus core protein Y132A mutant in complex with sulfamoylbenzamide (SBA_R01) | | Descriptor: | 4-fluoranyl-3-(4-oxidanylpiperidin-1-yl)sulfonyl-~{N}-[3,4,5-tris(fluoranyl)phenyl]benzamide, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
5WRE
 
 | | Hepatitis B virus core protein Y132A mutant in complex with heteroaryldihydropyrimidine (HAP_R01) | | Descriptor: | (2S)-1-[[(4R)-4-(2-chloranyl-4-fluoranyl-phenyl)-5-methoxycarbonyl-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-4,4-bis(fluoranyl)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-12-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
4W7T
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
8G0M
 
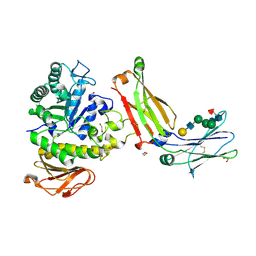 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
3FKY
 
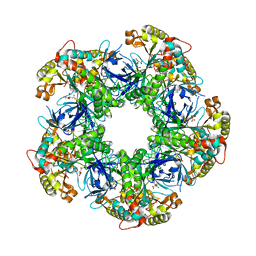 | | Crystal structure of the glutamine synthetase Gln1deltaN18 from the yeast Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, Glutamine synthetase | | Authors: | He, Y.X, Gui, L, Liu, Y.Z, Du, Y, Zhou, Y.Y, Li, P, Zhou, C.Z. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae glutamine synthetase Gln1 suggests a nanotube-like supramolecular assembly
Proteins, 76, 2009
|
|
9JK1
 
 | | Crystal structure of CDK12/Cyclin K in complex with covalent inhibitor YJZ5118 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Huang, W.X, Zhang, P.J, Yang, J.Z, Ding, K. | | Deposit date: | 2024-09-14 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery of YJZ5118 : A Potent and Highly Selective Irreversible CDK12/13 Inhibitor with Synergistic Effects in Combination with Akt Inhibition.
J.Med.Chem., 68, 2025
|
|
9LNQ
 
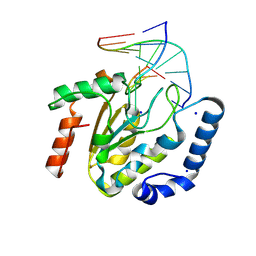 | | The hUNG bound to DNA product embedding 4primer-OCH3-dU | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(KBC)P*AP*TP*CP*TP*T)-3'), SODIUM ION, ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
9LNP
 
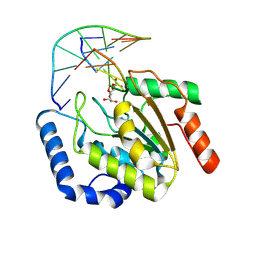 | | the hUNG bound to DNA product embedding uridine ribonucleotide | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*T*(RP5)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
6LG3
 
 | |
5KLI
 
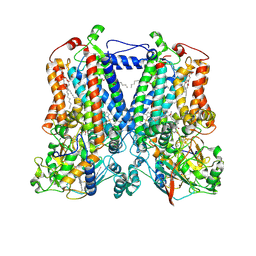 | | Rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5KKZ
 
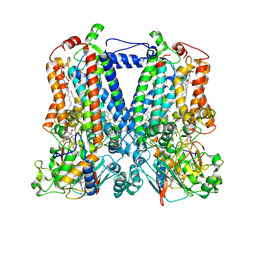 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5EWS
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-11-21 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
8YMB
 
 | | The crystal structure of SHD931 in complex with Brd4-BD2 and VCB | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, Elongin-B, ... | | Authors: | Huang, W.X, Chen, Y.H, Li, C.G, Ding, K. | | Deposit date: | 2024-03-08 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Based Design of "Head-to-Tail" Macrocyclic PROTACs.
Jacs Au, 4, 2024
|
|
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
3SR9
 
 | | Crystal structure of mouse PTPsigma | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Wang, J, Hou, L, Li, J, Ding, J. | | Deposit date: | 2011-07-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the homology and differences between mouse protein tyrosine phosphatase-sigma and human protein tyrosine phosphatase-sigma
Acta Biochim.Biophys.Sin., 43, 2011
|
|
