8EFP
 
 | |
8ET1
 
 | |
5GQD
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
7M8J
 
 | | SARS-CoV-2 S-NTD + Fab CM25 | | Descriptor: | CM25 Fab - Heavy Chain, CM25 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes.
Science, 372, 2021
|
|
5WZN
 
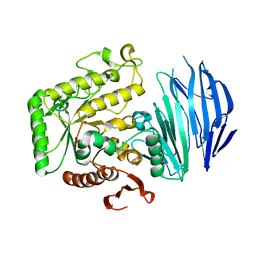 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
1BXE
 
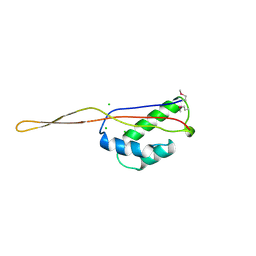 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
1LMC
 
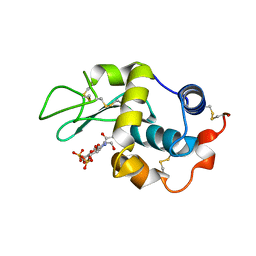 | | THE CRYSTAL STRUCTURE OF A COMPLEX BETWEEN BULGECIN, A BACTERIAL METABOLITE, AND LYSOZYME FROM THE RAINBOW TROUT | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Hough, E. | | Deposit date: | 1994-11-14 | | Release date: | 1996-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a complex between bulgecin, a bacterial metabolite, and lysozyme from the rainbow trout.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1IQW
 
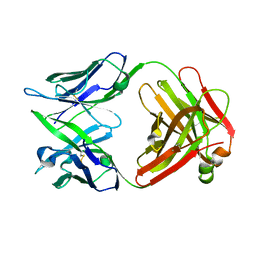 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
5LJJ
 
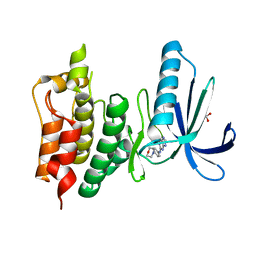 | | Crystal structure of human Mps1 (TTK) in complex with Reversine | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-07-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of reversine selectivity in inhibiting Mps1 more potently than aurora B kinase.
Proteins, 84, 2016
|
|
1LSP
 
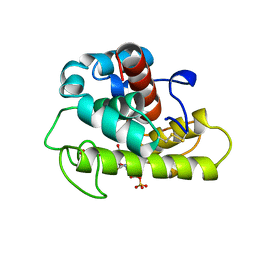 | | THE CRYSTAL STRUCTURE OF A BULGECIN-INHIBITED G-TYPE LYSOZYME FROM THE EGG-WHITE OF THE AUSTRALIAN BLACK SWAN. A COMPARISON OF THE BINDING OF BULGECIN TO THREE MURAMIDASES | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Rao, Z.H, Hough, E, Isaacs, N.W. | | Deposit date: | 1995-02-09 | | Release date: | 1996-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of a bulgecin-inhibited g-type lysozyme from the egg white of the Australian black swan. A comparison of the binding of bulgecin to three muramidases.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6IKN
 
 | | Crystal structure of the GAS7 F-BAR domain | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
6IKO
 
 | | Crystal structure of mouse GAS7cb | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.756 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
7D14
 
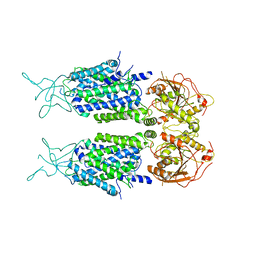 | | Mouse KCC2 | | Descriptor: | Solute carrier family 12 member 5 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
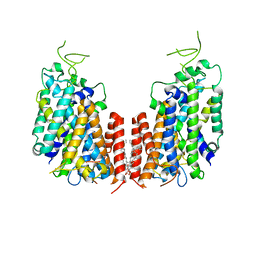 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
3W0Y
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, [3-fluoro-2'-methyl-4'-(3-{3-methyl-4-[(1E)-4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-en-1-yl]phenyl}pentan-3-yl)biphenyl-4-yl]acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure Analysis of Vitamin D receptor
TO BE PUBLISHED
|
|
2D20
 
 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D1Z
 
 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
5JZD
 
 | | A re-refinement of the isochorismate synthase EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
