1VZ3
 
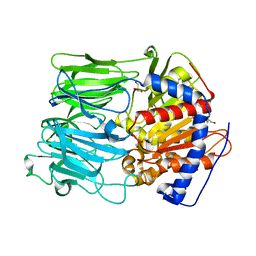 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, T597C MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Rea, D, Fulop, V. | | Deposit date: | 2004-05-14 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Concerted Structural Changes in the Peptidase and the Propeller Domains of Prolyl Oligopeptidase are Required for Substrate Binding
J.Mol.Biol., 340, 2004
|
|
2OM6
 
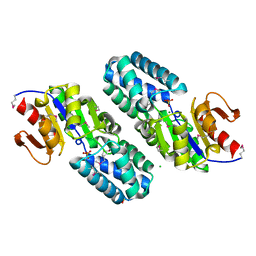 | | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable phosphoserine phosphatase, ... | | Authors: | Jeyakanthan, J, Vaijayanthimala, S, Gayathri, D, Velmurugan, D, Baba, S, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3
To be Published
|
|
6VRL
 
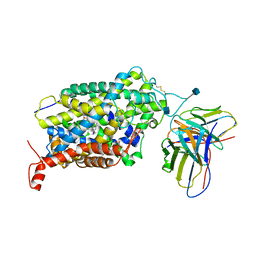 | | Cryo-EM structure of the wild-type human serotonin transporter complexed with I-paroxetine and 8B6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Navratna, V, Yang, D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chemical and structural investigation of the paroxetine-human serotonin transporter complex.
Elife, 9, 2020
|
|
7P9Q
 
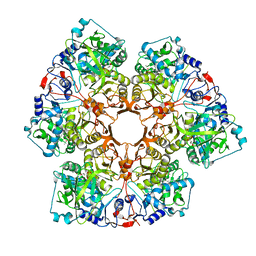 | | Crystal structure of Indole 3-Carboxylic acid decarboxylase from Arthrobacter nicotianae FI1612 in complex with co-factor prFMN. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, AnInD, MANGANESE (II) ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and biochemical characterization of the prenylated flavin mononucleotide-dependent indole-3-carboxylic acid decarboxylase.
J.Biol.Chem., 298, 2022
|
|
6FKB
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS13 | | Descriptor: | 2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK6
 
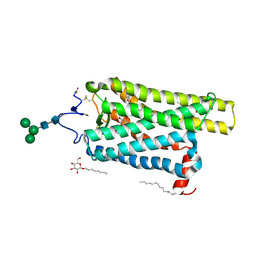 | | Crystal structure of N2C/D282C stabilized opsin bound to RS01 | | Descriptor: | (2~{S})-2-(4-chlorophenyl)-3-methyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6W32
 
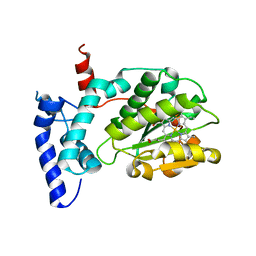 | | Crystal structure of Sfh5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phosphatidylinositol transfer protein SFH5 | | Authors: | Gulten, G, Khan, D, Aggarwal, A, Krieger, I, Sacchettini, J.C, Bankaitis, V.A. | | Deposit date: | 2020-03-08 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Sec14-like phosphatidylinositol transfer protein paralog defines a novel class of heme-binding proteins.
Elife, 9, 2020
|
|
7UHB
 
 | |
6FK8
 
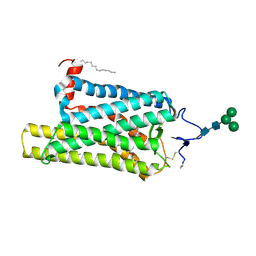 | | Crystal structure of N2C/D282C stabilized opsin bound to RS08 | | Descriptor: | (2~{R},3~{S})-3-azanyl-2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK7
 
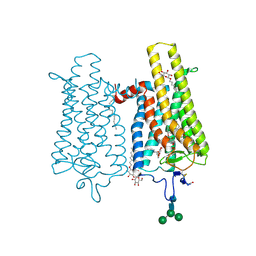 | | Crystal structure of N2C/D282C stabilized opsin bound to RS06 | | Descriptor: | (2~{R},3~{R})-2-(4-chlorophenyl)-3-oxidanyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FKD
 
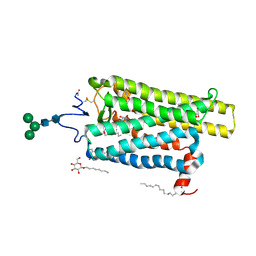 | | Crystal structure of N2C/D282C stabilized opsin bound to RS16 | | Descriptor: | 5-chloranyl-2-(2-oxidanylidene-2-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethyl)-3~{H}-pyridin-6-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6W2C
 
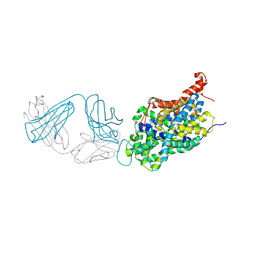 | | Anomalous iodine signal reveals the position of I-paroxetine complexed with the serotonin transporter at the central site | | Descriptor: | 8B6 heavy chain antibody fragment, 8B6 light chain antibody fragment, I-paroxetine, ... | | Authors: | Coleman, J.A, Navratna, V, Yang, D. | | Deposit date: | 2020-03-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (6.3 Å) | | Cite: | Chemical and structural investigation of the paroxetine-human serotonin transporter complex.
Elife, 9, 2020
|
|
1W9M
 
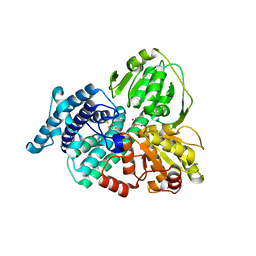 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6BQR
 
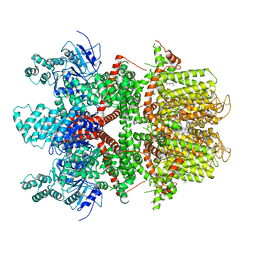 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
9DGO
 
 | |
6BQV
 
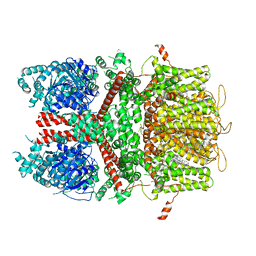 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-bound state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
6WZN
 
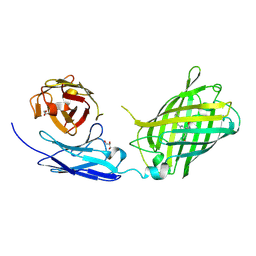 | | Crystal Structure of a Fluorescent Single Chain Fv Chimera | | Descriptor: | Fluorescent Single Chain Fv Chimera, GLYCEROL | | Authors: | Close, D, Velappan, N, Hung, L.W, Naranjo, L, Hemez, C, DeVore, N, McCullough, D, Lillo, A.M, Waldo, G, Bradbury, A.R.M. | | Deposit date: | 2020-05-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Construction, characterization and crystal structure of a fluorescent single-chain Fv chimera.
Protein Eng.Des.Sel., 34, 2021
|
|
2WBS
 
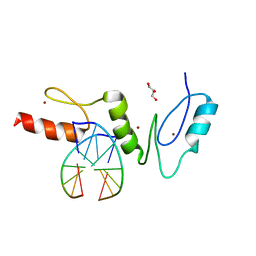 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | Descriptor: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | Authors: | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
7K4N
 
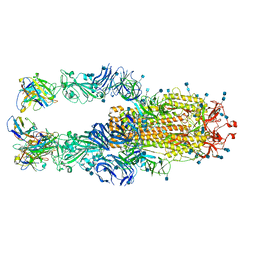 | |
6SI8
 
 | | Escherichia coli AGPase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6C2Q
 
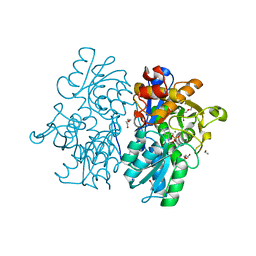 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-L-Serine Intermediate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
3BCM
 
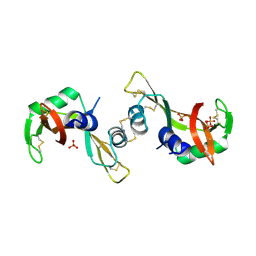 | | Crystal Structure of The Unswapped Form of P19A/L28Q/N67D BS-RNase | | Descriptor: | PHOSPHATE ION, Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
7QTK
 
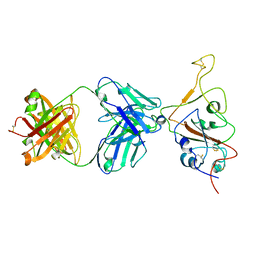 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTI
 
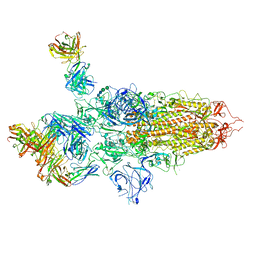 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTJ
 
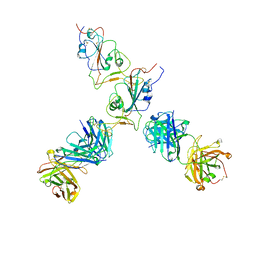 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
