5JGD
 
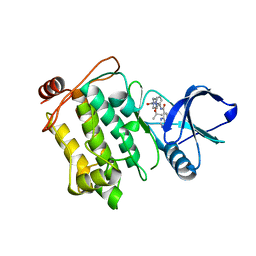 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 12 | | Descriptor: | N-(2-isopropoxy-3-(4-methylpiperazine-1-carbonyl)phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
8H79
 
 | |
7QGB
 
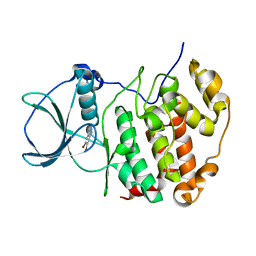 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 6.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, Casein kinase II subunit alpha | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
6X2I
 
 | |
6QBH
 
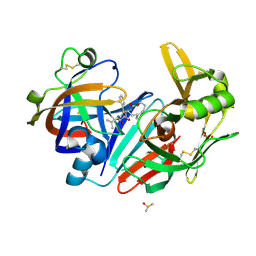 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 33 | | Descriptor: | (4~{S},5~{S},9~{S})-5-oxidanyl-4-(phenylmethyl)-9-propan-2-yl-1-oxa-3,8,11-triazacyclodocosane-2,7,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6QCB
 
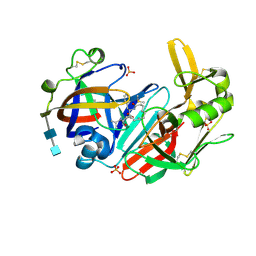 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 9 | | Descriptor: | (3~{S},7~{S},8~{S})-7-oxidanyl-8-(phenylmethyl)-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin D, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6OBA
 
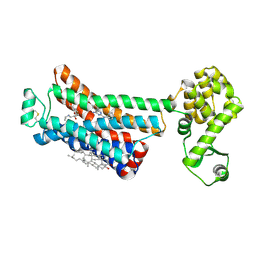 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
6OFI
 
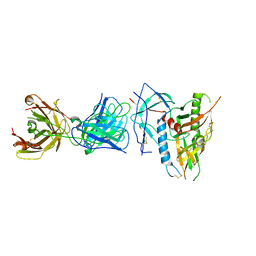 | | CRYSTAL STRUCTURE OF the RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH55 Fab heavy chain, CH55 Fab light chain, ... | | Authors: | Tolbert, W.D, Yan, F, Van, V, Pazgier, M. | | Deposit date: | 2019-03-29 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6QKY
 
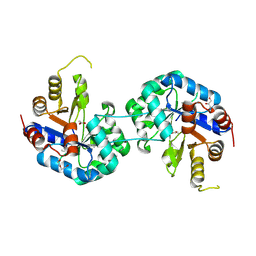 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8FYG
 
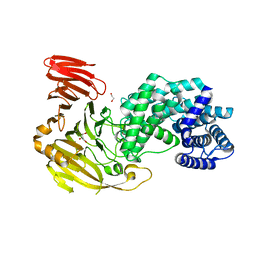 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8FNX
 
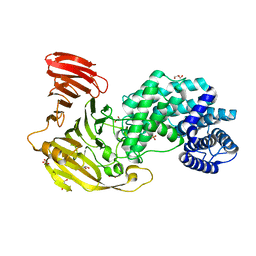 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8G0O
 
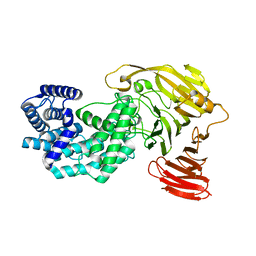 | | Crystal structure of Y281F mutant of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
6X2K
 
 | |
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0U
 
 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6OED
 
 | | CRYSTAL STRUCTURE OF THE RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB | | Descriptor: | CH55 Fab heavy chain, CH55 Fab light chain | | Authors: | Yan, F, Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6NXE
 
 | |
6WJ5
 
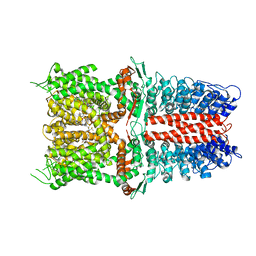 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
7UG7
 
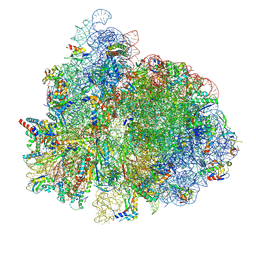 | | 70S ribosome complex in an intermediate state of translocation bound to EF-G(GDP) stalled by Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Wieland, M, Holm, M, Koller, T.O, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7A5M
 
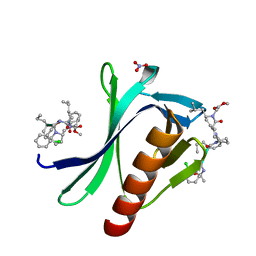 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe | | Descriptor: | Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6OY4
 
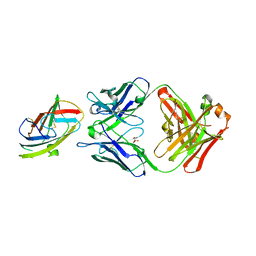 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
