7KHJ
 
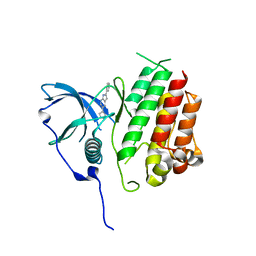 | | Crystal structure of KIT kinase domain with a small molecule inhibitor, PLX8512 in the DFG-in state | | Descriptor: | 2-phenyl-5-(1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Mast/stem cell growth factor receptor Kit | | Authors: | Zhang, Y. | | Deposit date: | 2020-10-21 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Association of Combination of Conformation-Specific KIT Inhibitors With Clinical Benefit in Patients With Refractory Gastrointestinal Stromal Tumors: A Phase 1b/2a Nonrandomized Clinical Trial.
Jama Oncol, 7, 2021
|
|
7KHK
 
 | |
7KHG
 
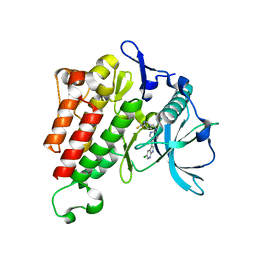 | | Crystal structure of KIT kinase domain with a small molecule inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Mast/stem cell growth factor receptor Kit | | Authors: | Zhang, Y. | | Deposit date: | 2020-10-21 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Association of Combination of Conformation-Specific KIT Inhibitors With Clinical Benefit in Patients With Refractory Gastrointestinal Stromal Tumors: A Phase 1b/2a Nonrandomized Clinical Trial.
Jama Oncol, 7, 2021
|
|
5LS6
 
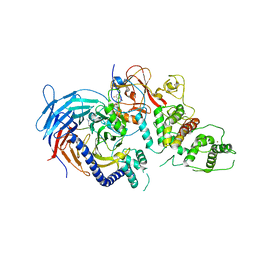 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | Descriptor: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
4K7F
 
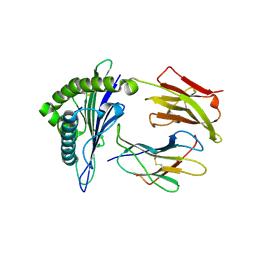 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
8AEY
 
 | | 3 A CRYO-EM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS FERRITIN FROM TIMEPIX3 detector | | Descriptor: | Ferritin BfrB | | Authors: | Zhang, Y, van Schayck, J.P, Knoops, K, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Integration of an Event-driven Timepix3 Hybrid Pixel Detector into a Cryo-EM Workflow
Microsc Microanal, 2023
|
|
1R0V
 
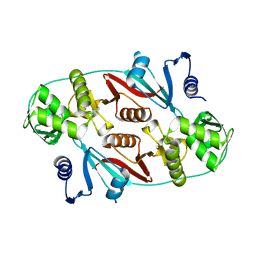 | |
7D6F
 
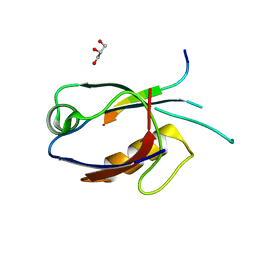 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
1R11
 
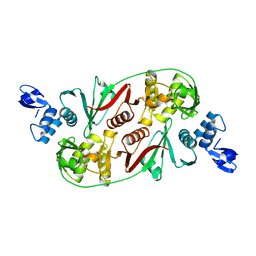 | |
3LZD
 
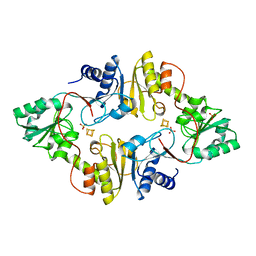 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
4F6X
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1112 | | Descriptor: | 1-[3-(hexyloxy)benzyl]-4-hydroxy-2-oxo-1,2-dihydropyridine-3-carboxylic acid, D(-)-TARTARIC ACID, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
4MQ9
 
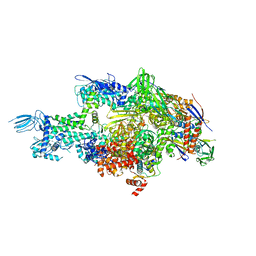 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4F6V
 
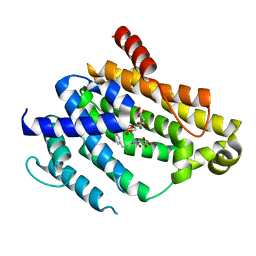 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1034, mg2+ and fmp. | | Descriptor: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 2,4-dioxo-4-{[3-(3-phenoxyphenyl)propyl]amino}butanoic acid, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
6C00
 
 | |
6WXK
 
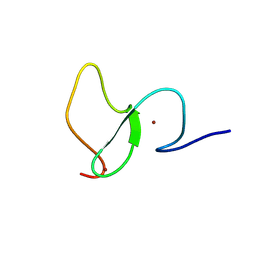 | | PHF23 PHD Domain Apo | | Descriptor: | PHD finger protein 23, ZINC ION | | Authors: | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
3NTP
 
 | | Human Pin1 complexed with reduced amide inhibitor | | Descriptor: | (2R)-2-(acetylamino)-3-[(2S)-2-{[2-(1H-indol-3-yl)ethyl]carbamoyl}pyrrolidin-1-yl]propyl dihydrogen phosphate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, Y. | | Deposit date: | 2010-07-05 | | Release date: | 2012-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A reduced-amide inhibitor of Pin1 binds in a conformation resembling a twisted-amide transition state.
Biochemistry, 50, 2011
|
|
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
5WTB
 
 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
3PWM
 
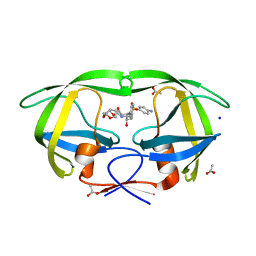 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3RY0
 
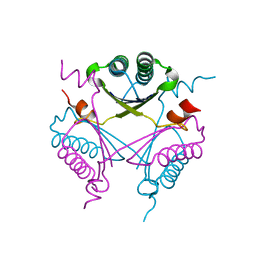 | | Crystal structure of TomN, a 4-Oxalocrotonate Tautomerase homologue in Tomaymycin biosynthetic pathway | | Descriptor: | Putative tautomerase | | Authors: | Zhang, Y, Yan, W.P, Li, W.Z, Whitman, C.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Biochemistry, 50, 2011
|
|
5Y4O
 
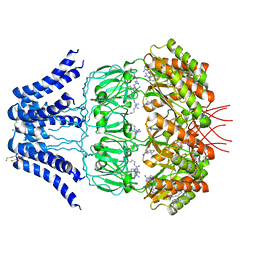 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
7T1F
 
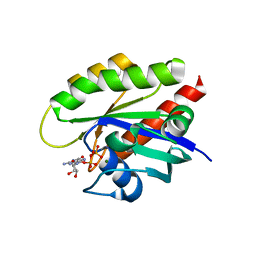 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
4GA3
 
 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1260 | | Descriptor: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2012-07-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemo-Immunotherapeutic Anti-Malarials Targeting Isoprenoid Biosynthesis.
ACS MED.CHEM.LETT., 4, 2013
|
|
3CIV
 
 | |
