8TZ4
 
 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, Sodium/nucleoside cotransporter | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 20, 2024
|
|
8TZ2
 
 | |
8TZD
 
 | |
8TZ1
 
 | | Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, ... | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Antiviral drug recognition and elevator-type transport motions of CNT3.
Nat.Chem.Biol., 20, 2024
|
|
8TZ7
 
 | |
8TZ5
 
 | |
8TZ6
 
 | |
8TZA
 
 | |
8TZ3
 
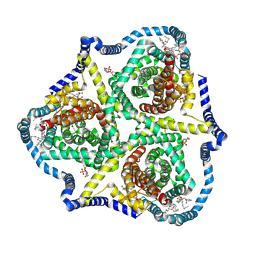 | |
8TZ8
 
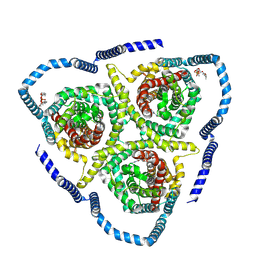 | |
8TZ9
 
 | |
3SWD
 
 | | E. coli MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys115 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
6LNA
 
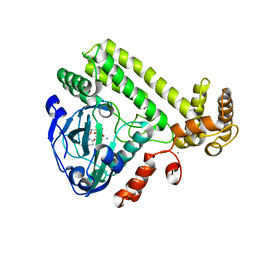 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
7Y7J
 
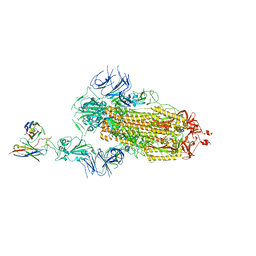 | | SARS-CoV-2 S trimer in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, Spike protein S1 | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7BYM
 
 | | Cryo-EM structure of human KCNQ4 with retigabine | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYL
 
 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
7BYN
 
 | | Cryo-EM structure of human KCNQ4 with linopirdine | | Descriptor: | 1-phenyl-3,3-bis(pyridin-4-ylmethyl)indol-2-one, Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
5UE5
 
 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
5UE2
 
 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
3DK3
 
 | |
5VY6
 
 | | A self-assembling D-form DNA crystal lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, Yan, H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | Tuning the Cavity Size and Chirality of Self-Assembling 3D DNA Crystals.
J. Am. Chem. Soc., 139, 2017
|
|
5VY7
 
 | | A self-assembling L-form DNA crystal lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, Yan, H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tuning the Cavity Size and Chirality of Self-Assembling 3D DNA Crystals.
J. Am. Chem. Soc., 139, 2017
|
|
3DK6
 
 | |
3DK7
 
 | |
