1QB6
 
 | | BOVINE TRYPSIN 3,3'-[3,5-DIFLUORO-4-METHYL-2, 6-PYRIDINEDIYLBIS(OXY)]BIS(BENZENECARBOXIMIDAMIDE) (ZK-805623) COMPLEX | | Descriptor: | 3,3'-[3,5-DIFLUORO-4-METHYL-2,6-PYRIDYLENEBIS(OXY)]-BIS(BENZENECARBOXIMIDAMIDE), CALCIUM ION, POTASSIUM ION, ... | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6IC9
 
 | |
6IC8
 
 | |
4LIQ
 
 | | Structure of the extracellular domain of human CSF-1 receptor in complex with the Fab fragment of RG7155 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment RG7155 heavy chain, Fab fragment RG7155 light chain, ... | | Authors: | Benz, J, Gorr, I.H, Hertenberger, H, Ries, C.H. | | Deposit date: | 2013-07-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy
Cancer Cell, 25, 2014
|
|
6F9M
 
 | | The LIPY/F-motif in an intracellular subtilisin protease is involved in inhibition | | Descriptor: | ACETATE ION, SODIUM ION, Serine protease, ... | | Authors: | Bjerga, G.E.K, Larsen, O, Arsin, H, Williamson, A.K, Garcia-Moyano, A, Leiros, I, Puntervoll, P. | | Deposit date: | 2017-12-14 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mutational analysis of the pro-peptide of a marine intracellular subtilisin protease supports its role in inhibition.
Proteins, 86, 2018
|
|
4L9L
 
 | |
1QBO
 
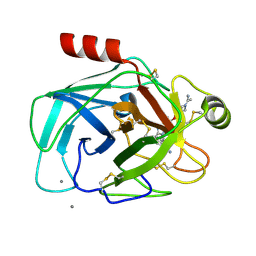 | |
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8VW5
 
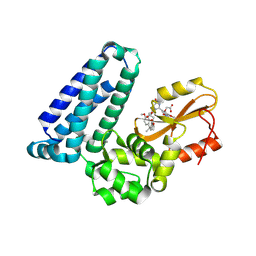 | | Crystal structure of Cbl-b TKB bound to compound 2 | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL-B, MAGNESIUM ION, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
8VW4
 
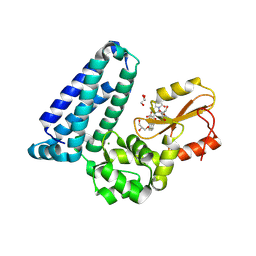 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
3L1A
 
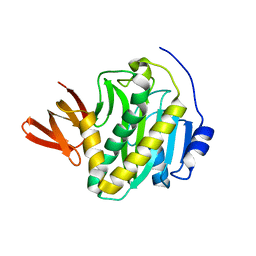 | |
7B3E
 
 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
8R0I
 
 | | Pseudomonas aeruginosa FabF C164A in complex with 3-amino-N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-N-(1,5-dimethyl-3-oxidanylidene-2-phenyl-pyrazol-4-yl)benzamide, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8R1V
 
 | | Pseudomonas aeruginosa FabF C164A in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-(4-methoxyphenoxy)acetamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
7ALI
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7OND
 
 | |
7OO4
 
 | |
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, TRYPSIN | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-09 | | Release date: | 2000-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6Q2V
 
 | |
6Q5Y
 
 | |
7SKI
 
 | | Pertussis toxin in complex with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.09912443 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKY
 
 | | Pertussis toxin S1 bound to NAD+ | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37000132 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7SKK
 
 | | pertussis toxin in complex with ADPR and Nicotinamide | | Descriptor: | NICOTINAMIDE, Pertussis toxin subunit 1, SULFATE ION, ... | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65000772 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8B4N
 
 | |
