4XNW
 
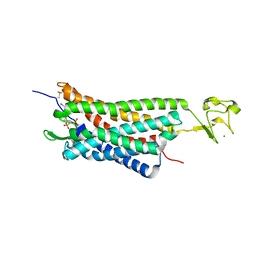 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4XNV
 
 | | The human P2Y1 receptor in complex with BPTU | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2-(2-tert-butylphenoxy)pyridin-3-yl]-3-[4-(trifluoromethoxy)phenyl]urea, CHOLESTEROL, ... | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4YWZ
 
 | |
2YYF
 
 | | Purification and structural characterization of a D-amino acid containing conopeptide, marmophine, from Conus marmoreus | | Descriptor: | M-conotoxin mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C. | | Deposit date: | 2007-04-29 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a D-amino acid-containing conopeptide, conomarphin, from Conus marmoreus.
Febs J., 275, 2008
|
|
6M2W
 
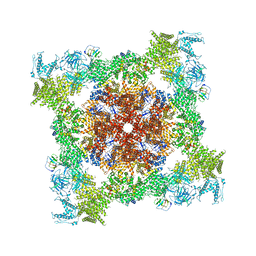 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6V9T
 
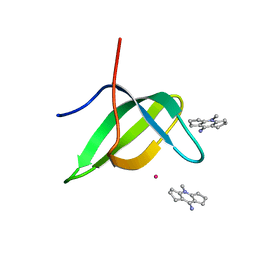 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
8HDO
 
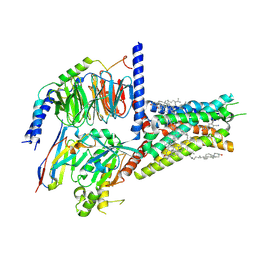 | | Structure of A2BR bound to synthetic agonists BAY 60-6583 | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
8HDP
 
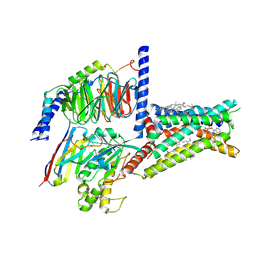 | | Structure of A2BR bound to endogenous agonists adenosine | | Descriptor: | ADENOSINE, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
5GON
 
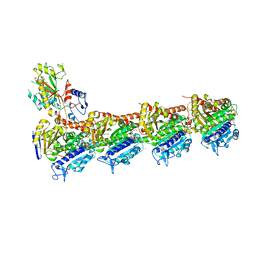 | | Structures of a beta-lactam bridged analogue in complex with tubulin | | Descriptor: | (3R,4R)-4-(4-methoxy-3-oxidanyl-phenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhou, L, Liu, Y, Cheng, L, Wang, Y. | | Deposit date: | 2016-07-28 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Potent Antitumor Activities and Structure Basis of the Chiral beta-Lactam Bridged Analogue of Combretastatin A-4 Binding to Tubulin.
J. Med. Chem., 59, 2016
|
|
5TJ8
 
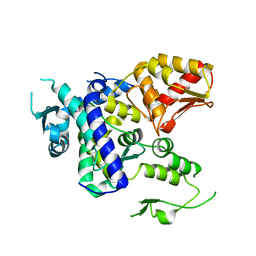 | |
5TJ7
 
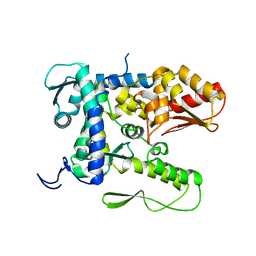 | |
5TJQ
 
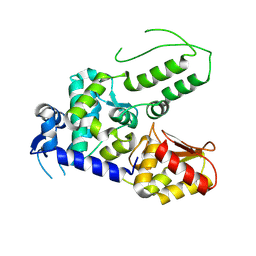 | | Structure of WWP2 2,3-linker-HECT | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2,NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2016-10-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Tunable Brake for HECT Ubiquitin Ligases.
Mol. Cell, 66, 2017
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
5BU3
 
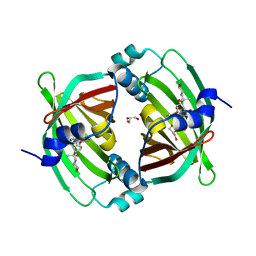 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
8H8J
 
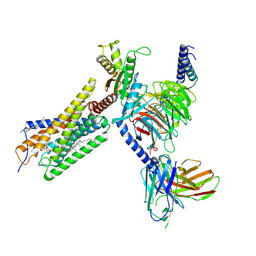 | | Lodoxamide-bound GPR35 in complex with G13 | | Descriptor: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | Authors: | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-10-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
8HBD
 
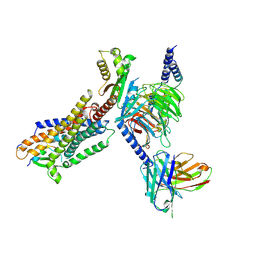 | | Cryo-EM structure of IRL1620-bound ETBR-Gi complex | | Descriptor: | Endothelin receptor type B,Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Ji, Y, Jiang, Y, Duan, J, Xu, H.E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
8HCX
 
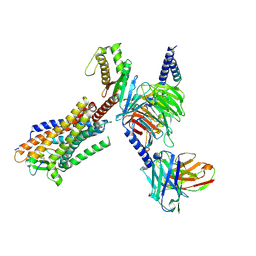 | | Cryo-EM structure of Endothelin1-bound ETBR-Gq complex | | Descriptor: | Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
8HCQ
 
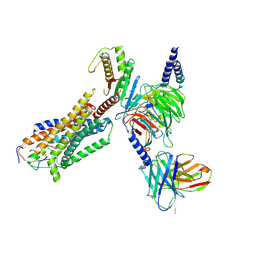 | | Cryo-EM structure of endothelin1-bound ETAR-Gq complex | | Descriptor: | Endothelin-1, Endothelin-1 receptor,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
