6X1U
 
 | | Structure of pHis Fab (SC39-4) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACLYana-3-pTza peptide, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4QGT
 
 | | The Crystal Structure of Human IgG Fc Domain with Enhanced Aromatic Sequon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kong, L, Connelly, S.C, Wilson, I.A. | | Deposit date: | 2014-05-25 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Stabilizing the CH2 Domain of an Antibody by Engineering in an Enhanced Aromatic Sequon.
Acs Chem.Biol., 11, 2016
|
|
6X1S
 
 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6OPA
 
 | |
6XKQ
 
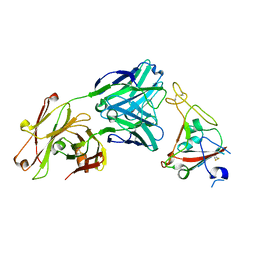 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
4R26
 
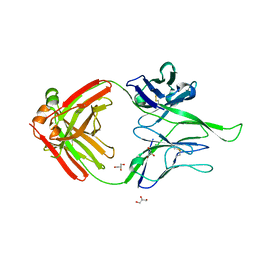 | | Crystal structure of human Fab PGT124, a broadly neutralizing and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGR124-Light Chain, PGT124-Heavy Chain | | Authors: | Garces, F, Kong, L, Wilson, I.A. | | Deposit date: | 2014-08-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4969 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
4R2G
 
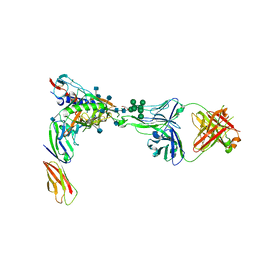 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
6PBW
 
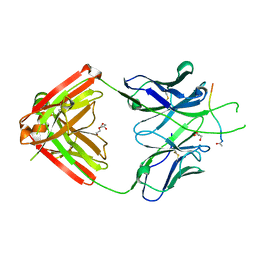 | | Crystal structure of Fab667 complex | | Descriptor: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
6PBV
 
 | | Crystal structure of Fab668 complex | | Descriptor: | 1,2-ETHANEDIOL, Fab668 heavy chain, Fab668 light chain, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.566 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
4RQQ
 
 | |
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6PZD
 
 | |
6Q1Z
 
 | |
4RNR
 
 | | Crystal structure of broadly neutralizing anti-HIV antibody PGT130 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PGT130 Heavy Chain, PGT130 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Two Classes of Broadly Neutralizing Antibodies within a Single Lineage Directed to the High-Mannose Patch of HIV Envelope.
J.Virol., 89, 2015
|
|
4RQS
 
 | | Crystal structure of fully glycosylated HIV-1 gp120 core bound to CD4 and 17b Fab | | Descriptor: | 17b Fab Heavy Chain, 17b Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A, Kwong, P.D. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.493 Å) | | Cite: | Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262.
Proteins, 83, 2015
|
|
6PZF
 
 | |
6PZE
 
 | |
6WOS
 
 | |
6O42
 
 | |
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6PZH
 
 | | Crystal structure of human NA-22 Fab | | Descriptor: | NA-22 Fab heavy chain, NA-22 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6PZG
 
 | | Crystal structure of human NA-80 Fab | | Descriptor: | NA-80 Fab heavy chain, NA-80 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6Q23
 
 | |
6Q20
 
 | |
