209D
 
 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
7K5I
 
 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
7Y6L
 
 | |
7Y6N
 
 | |
4W4O
 
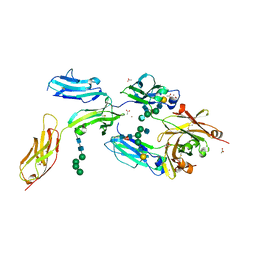 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
1LZ5
 
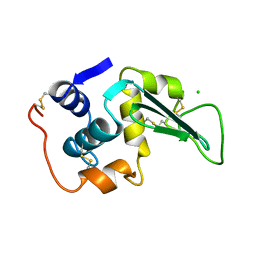 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
3KC0
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 10b | | Descriptor: | Fructose-1,6-bisphosphatase 1, [(8H-indeno[1,2-d][1,3]thiazol-4-yloxy)methyl]phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4WUA
 
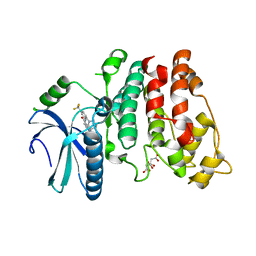 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | Descriptor: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | Authors: | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
7DKJ
 
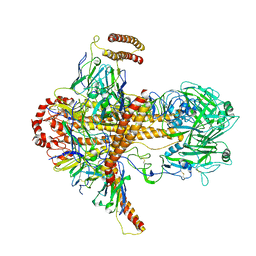 | |
5XI9
 
 | |
6VHF
 
 | | Crystal structure of RbBP5 interacting domain of Cfp1 | | Descriptor: | PHD-type domain-containing protein, ZINC ION | | Authors: | Joshi, M, Couture, J.F. | | Deposit date: | 2020-01-09 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | A non-canonical monovalent zinc finger stabilizes the integration of Cfp1 into the H3K4 methyltransferase complex COMPASS.
Nucleic Acids Res., 48, 2020
|
|
4W4N
 
 | | Crystal structure of human Fc at 1.80 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
6C3E
 
 | | CRYSTAL STRUCTURE OF RIP1 KINASE BOUND TO INHIBITOR | | Descriptor: | 2-benzyl-5-nitro-1H-benzimidazole, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
2RNE
 
 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
2RU3
 
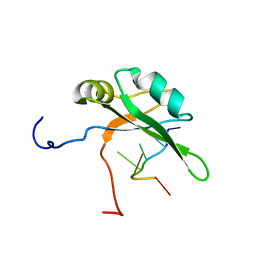 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
3D7A
 
 | |
6E2H
 
 | |
5B86
 
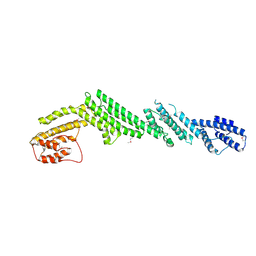 | | Crystal structure of M-Sec | | Descriptor: | Tumor necrosis factor alpha-induced protein 2 | | Authors: | Yamashita, M, Sato, Y, Yamagata, A, Fukai, S. | | Deposit date: | 2016-06-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.017 Å) | | Cite: | Distinct Roles for the N- and C-terminal Regions of M-Sec in Plasma Membrane Deformation during Tunneling Nanotube Formation.
Sci Rep, 6, 2016
|
|
5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNX
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNW
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
