6Y1X
 
 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
7BI7
 
 | | An Unexpected P-Cluster like Intermediate En Route to the Nitrogenase FeMo-co | | Descriptor: | FE(8)-S(7) CLUSTER, FeMo cofactor biosynthesis protein NifB, HYDROSULFURIC ACID, ... | | Authors: | Jenner, L.P, Cherrier, M.V, Amara, P, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An unexpected P-cluster like intermediate en route to the nitrogenase FeMo-co.
Chem Sci, 12, 2021
|
|
6FD2
 
 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
3T7V
 
 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3IIZ
 
 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with S-adenosyl-L-methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3IIX
 
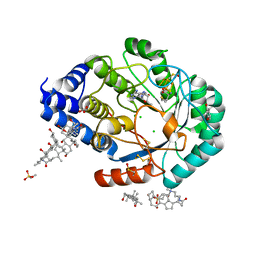 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with methionine and 5'deoxyadenosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBONATE ION, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
7JMA
 
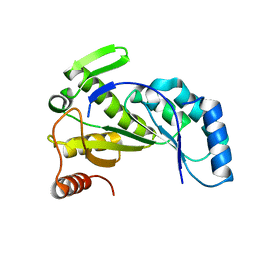 | |
7JMB
 
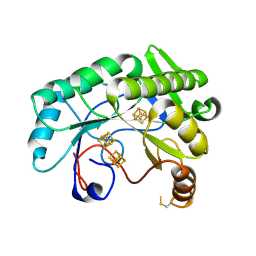 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | Authors: | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4U0P
 
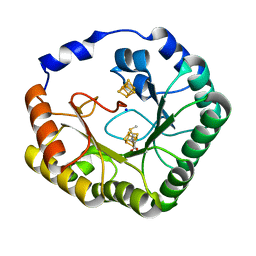 | | The Crystal Structure of Lipoyl Synthase in Complex with S-Adenosyl Homocysteine | | Descriptor: | IRON/SULFUR CLUSTER, Lipoyl synthase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Sandy, J, Dinis, P.C, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
4U0O
 
 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
5FES
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF2
 
 | | HydE from T. maritima in complex with (2R,4R)-TDA | | Descriptor: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF4
 
 | | HydE from T. maritima in complex with (2R,4R)-TMeTDA | | Descriptor: | (2~{R},4~{R})-2,5,5-trimethyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
3CAN
 
 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
5FEZ
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA, 5'-deoxyadenosine and methionine | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
3CIX
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima in complex with thiocyanate | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
7VOC
 
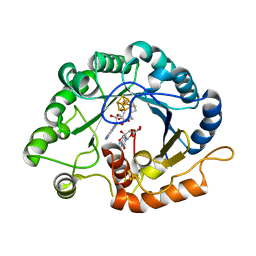 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
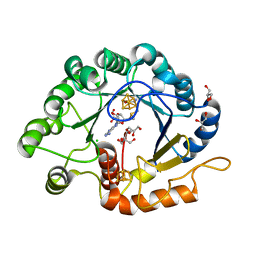 | |
7WZX
 
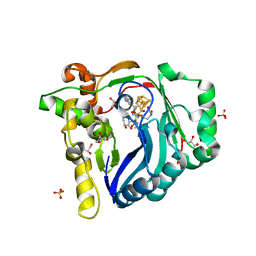 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
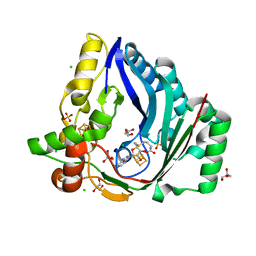 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
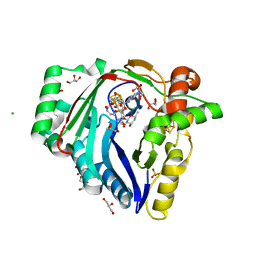 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7O25
 
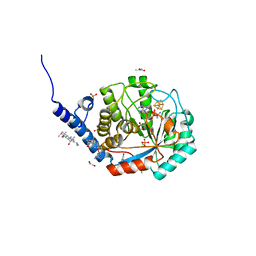 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1P
 
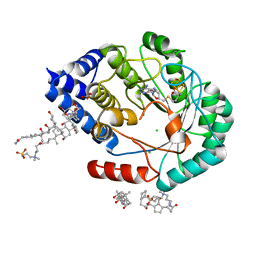 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
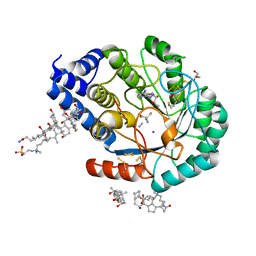 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
