3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVP
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (10-SKL) | | Descriptor: | 10-SKL PTS1 peptide Ac-GTLSNRASKL, Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CV0
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3ESK
 
 | |
3FP2
 
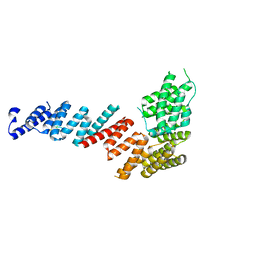 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP3
 
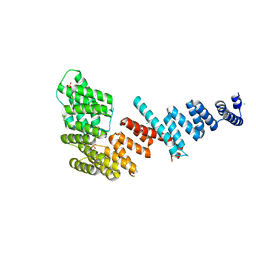 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP4
 
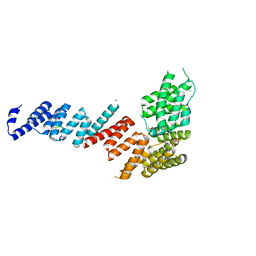 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
7Y4I
 
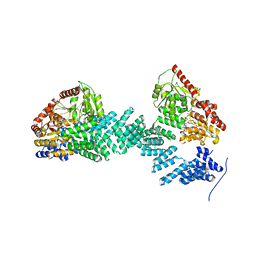 | | Crystal structure of SPINDLY in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Xu, S.T, Wan, L.H. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into mechanism and specificity of the plant protein O-fucosyltransferase SPINDLY.
Nat Commun, 13, 2022
|
|
7YEA
 
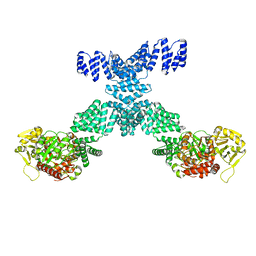 | | Human O-GlcNAc transferase Dimer | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gao, H, Lu, P, Liu, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
7YEH
 
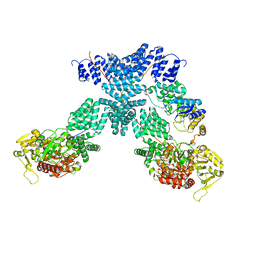 | | Cryo-EM structure of human OGT-OGA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Lu, P, Liu, Y, Yu, H, Gao, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
8A61
 
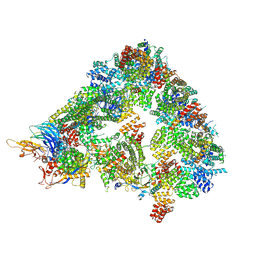 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A5Y
 
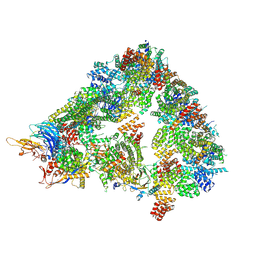 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
7QE7
 
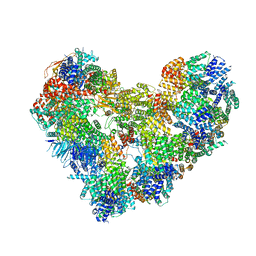 | | High-resolution structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
6Q6G
 
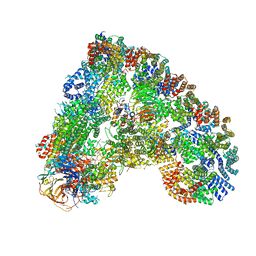 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
5A31
 
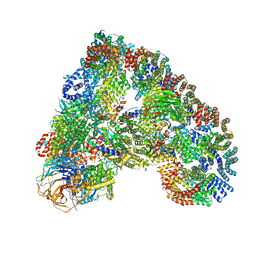 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
5OMP
 
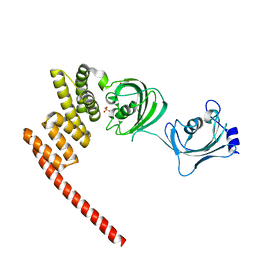 | | Human FKBP5 protein | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, SULFATE ION | | Authors: | Kumar, R, Moche, M, Winblad, B, Pavlov, P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Combined x-ray crystallography and computational modeling approach to investigate the Hsp90 C-terminal peptide binding to FKBP51.
Sci Rep, 7, 2017
|
|
1P5Q
 
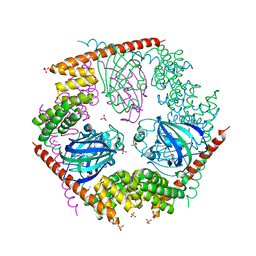 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7OBE
 
 | |
5JJT
 
 | |
5JHE
 
 | |
1WAO
 
 | | PP5 structure | | Descriptor: | MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Barford, D. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Tpr Domain-Mediated Regulation of Protein Phosphatase 5
Embo J., 24, 2005
|
|
1KT0
 
 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | 51 KDA FK506-BINDING PROTEIN, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a
component of steroid receptor complexes
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1KT1
 
 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | FK506-binding protein FKBP51, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURE OF THE LARGE FK506-BINDING PROTEIN FKBP51, AN HSP90-BINDING PROTEIN AND A COMPONENT OF STEROID RECEPTOR COMPLEXES
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QZ2
 
 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8S4G
 
 | | Cryo-EM structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2024-02-21 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | New structural features of the APC/C revealed by high resolution cryo-EM structures of apo-APC/C and the APC/C-CDH1-EMI1 complex
To Be Published
|
|
