1ZP2
 
 | |
1BU2
 
 | |
2PK2
 
 | | Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat | | Descriptor: | Cyclin-T1, Protein Tat | | Authors: | Anand, K, Schulte, A, Fujinaga, K, Scheffzek, K, Geyer, M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Cyclin Box Structure of the P-TEFb Subunit Cyclin T1 Derived from a Fusion Complex with EIAV Tat.
J.Mol.Biol., 370, 2007
|
|
1VIN
 
 | |
2B9R
 
 | |
1JKW
 
 | |
1KXU
 
 | | CYCLIN H, A POSITIVE REGULATORY SUBUNIT OF CDK ACTIVATING KINASE | | Descriptor: | CYCLIN H | | Authors: | Kim, K.K, Chamberin, H.M, Morgan, D.O, Kim, S.-H. | | Deposit date: | 1996-08-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase.
Nat.Struct.Biol., 3, 1996
|
|
2IVX
 
 | | Crystal structure of human cyclin T2 at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-T2 | | Authors: | Debreczeni, J.E, Bullock, A.N, Fedorov, O, Savitsky, P, Berridge, G, Das, S, Pike, A.C.W, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Gorrec, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation.
EMBO J., 27, 2008
|
|
2I53
 
 | | Crystal structure of Cyclin K | | Descriptor: | ACETATE ION, Cyclin K | | Authors: | Baek, K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2006-08-23 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Cyclin K, a Positive Regulator of Cyclin-dependent Kinase 9.
J.Mol.Biol., 366, 2007
|
|
6Q6H
 
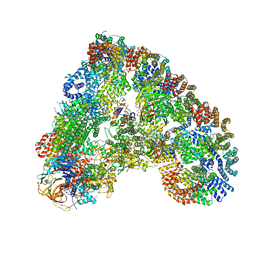 | |
7LUO
 
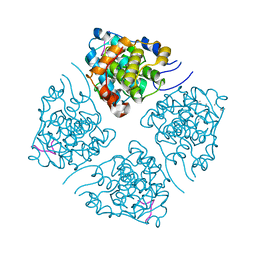 | | N-terminus of Skp2 bound to Cyclin A | | Descriptor: | S-phase kinase-associated protein 2,Cyclin-A2, Skp2 Motif 1 uncharacterized fragment 1, Skp2 Motif 1 uncharacterized fragment 2 | | Authors: | Kelso, S, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Bipartite binding of the N terminus of Skp2 to cyclin A.
Structure, 29, 2021
|
|
6Q6G
 
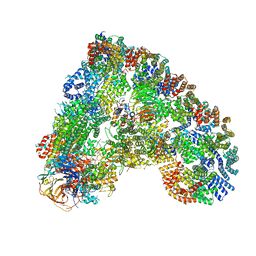 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
2W2H
 
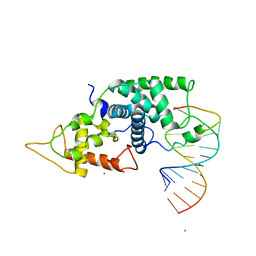 | |
7NWK
 
 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
3LQ5
 
 | | Structure of CDK9/CyclinT in complex with S-CR8 | | Descriptor: | (2S)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Hole, A.J, Endicott, J.A, Baumli, S. | | Deposit date: | 2010-02-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CDK Inhibitors Roscovitine and CR8 Trigger Mcl-1 Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells
Genes Cancer, 1, 2010
|
|
1E9H
 
 | | Thr 160 phosphorylated CDK2 - Human cyclin A3 complex with the inhibitor indirubin-5-sulphonate bound | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, CELL DIVISION PROTEIN KINASE 2, CYCLIN A3 | | Authors: | Davies, T.G, Tunnah, P, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2000-10-16 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor Binding to Active and Inactive Cdk2: The Crystal Structure of Cdk2-Cyclin A/Indirubin-5-Sulphonate
Structure, 9, 2001
|
|
8I0L
 
 | | Structure of CDK9/cyclin T1 in complex with inhibitor | | Descriptor: | 2-[(4-azanylcyclohexyl)amino]-7-cyclopentyl-~{N},~{N}-dimethyl-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Jiang, C, Ye, Y, Huang, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of CDK9/cyclin T1 in complex with inhibitor
To Be Published
|
|
4G6L
 
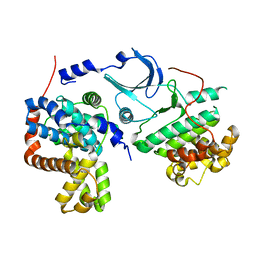 | | Crystal structure of human CDK8/CYCC in the DMG-in conformation | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID | | Authors: | Schneider, E.V, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2012-07-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4UN0
 
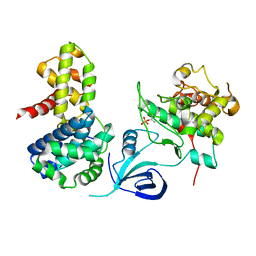 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
3DDQ
 
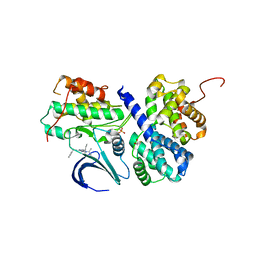 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor roscovitine | | Descriptor: | Cell division protein kinase 2, Cyclin-A2, MONOTHIOGLYCEROL, ... | | Authors: | Echalier, A, Endicott, J.A. | | Deposit date: | 2008-06-06 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CR8, a potent and selective, roscovitine-derived inhibitor of cyclin-dependent kinases.
Oncogene, 27, 2008
|
|
5ACB
 
 | | Crystal Structure of the Human Cdk12-Cyclink Complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Mackenzie, A, Goubin, S, Strain-Damerell, C, Mahajan, P, Tallant, C, Chalk, R, Wiggers, H, Kopec, J, Fitzpatrick, F, Burgess-Brown, N, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-15 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Targeting of Remote Cysteine Residues to Develop Cdk12 and Cdk13 Inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
2UUE
 
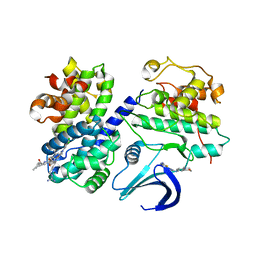 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | 1-(3,5-DICHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOLE-3-CARBOXYLIC ACID, 4-METHYL-5-{(2E)-2-[(4-MORPHOLIN-4-YLPHENYL)IMINO]-2,5-DIHYDROPYRIMIDIN-4-YL}-1,3-THIAZOL-2-AMINE, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-03-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
7XQK
 
 | | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, G1/S-specific cyclin-E1, GLYCEROL, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex from Biortus.
To Be Published
|
|
6O9L
 
 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4TTH
 
 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
