2MSL
 
 | |
2MSN
 
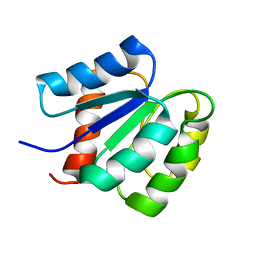
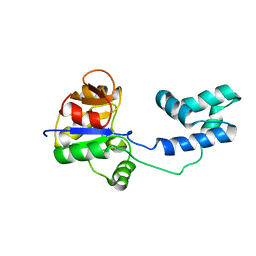 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MSO
 
 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSQ
 
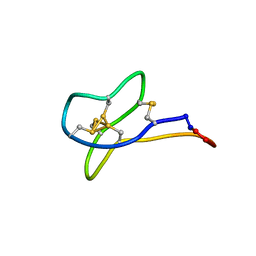
 | | Solution study of cBru9a | | Descriptor: | Conotoxin cBru9a | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSR
 
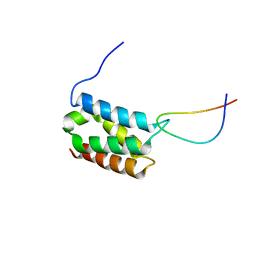 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
2MSU
 
 | |
2MSV
 
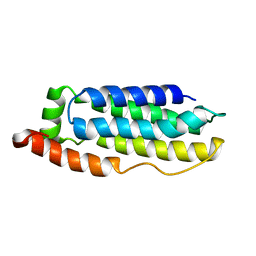 | | Solution structure of the MLKL N-terminal domain | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Su, L, Rizo, J, Quade, B, Wang, H, Sun, L, Wang, X. | | Deposit date: | 2014-08-07 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Plug Release Mechanism for Membrane Permeation by MLKL.
Structure, 22, 2014
|
|
2MSW
 
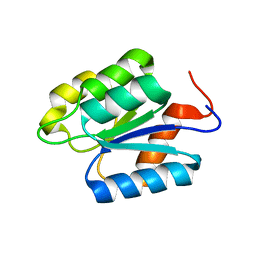 | |
2MSX
 
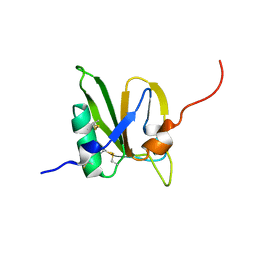 | | The solution structure of the MANEC-type domain from Hepatocyte Growth Factor Inhibitor 1 reveals an unexpected PAN/apple domain-type fold | | Descriptor: | Kunitz-type protease inhibitor 1 | | Authors: | Hong, Z, Nowakowski, M.E, Spronk, C, Petersen, S.V, Petersen, J.S, Kozminski, W, Mulder, F, Jensen, J.K. | | Deposit date: | 2014-08-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Biochem.J., 466, 2015
|
|
2MSY
 
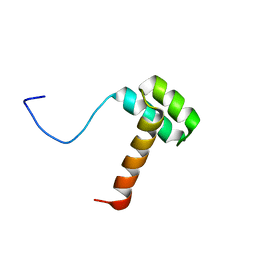 | | Solution structure of Hox homeodomain | | Descriptor: | Homeobox protein Hox-C9 | | Authors: | Kim, H, Park, S, Han, J, Lee, B. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction between the Hox and HMGB1 and understanding of the HMGB1-enhancing effect of Hox-DNA binding.
Biochim.Biophys.Acta, 1854, 2015
|
|
2MT3
 
 | |
2MT4
 
 | |
2MT5
 
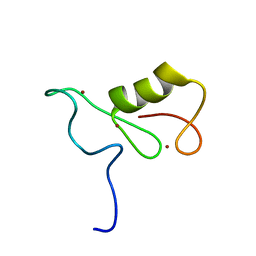 | | Isolated Ring domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissman, F, Royappa, G, Schulman, B, Jarvis, M, Vanderlinden, R, Frye, J.J, Qiao, R, Petzold, G, Peters, J, Stark, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
2MT6
 
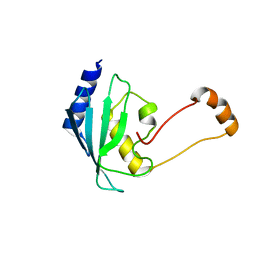 | | Solution structure of the human ubiquitin conjugating enzyme Ube2w | | Descriptor: | Ubiquitin-conjugating enzyme E2 W | | Authors: | Vittal, V, Shi, L, Wenzel, D.M, Brzovic, P.S, Klevit, R.E. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Nat.Chem.Biol., 11, 2015
|
|
2MT7
 
 | |
2MT8
 
 | |
2MT9
 
 | | Solution structure of holo_FldB | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin-2 | | Authors: | Jin, C, Fu, W, Ye, Q. | | Deposit date: | 2014-08-15 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Long-chain flavodoxin FldB from Escherichia coli
J.Biomol.Nmr, 60, 2014
|
|
2MTB
 
 | | Solution structure of apo_FldB | | Descriptor: | Flavodoxin-2 | | Authors: | Jin, C, Fu, W, Ye, Q. | | Deposit date: | 2014-08-15 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Long-chain flavodoxin FldB from Escherichia coli
J.Biomol.Nmr, 60, 2014
|
|
2MTC
 
 | |
2MTD
 
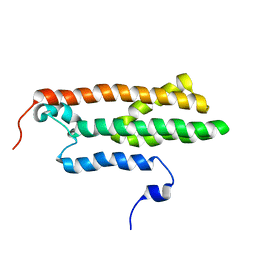 | |
2MTE
 
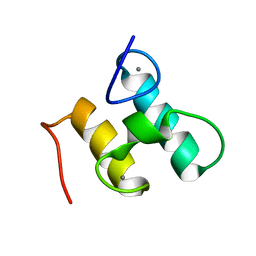 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTG
 
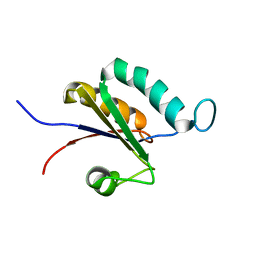 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTI
 
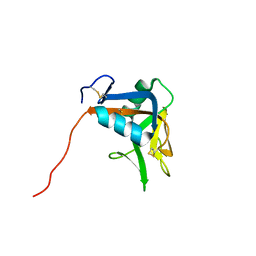 | | NMR structure of the lymphocyte receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A | | Authors: | Chmelik, J, Rozbesky, D, Pospisilova, E, Adamek, D, Novak, P. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lymphocyte receptor Nkrp1a reveals a distinct conformation of the long loop region as compared to in the crystal structure.
Proteins, 84, 2016
|
|
2MTJ
 
 | |
